
[ad_1]
Genome research conducted by the University of Warwick suggests that enteric fever, a potentially life-threatening disease more prevalent in hot countries, was present in medieval Europe.
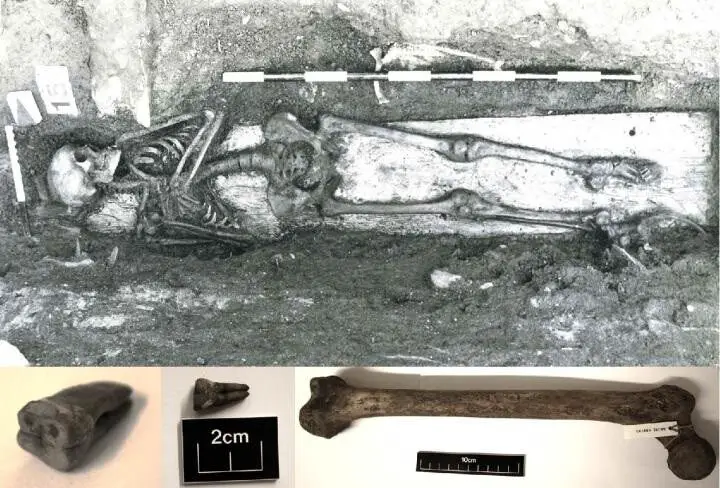
Salmonella Paratyphi C A potentially fatal infection was detected in an 800-year-old human skeleton discovered in Trondheim, Norway
Scientists now speculate that the evolution of Enteric fever may be linked to the domestication of pigs in Northern Europe.
The research was conducted by a team of international collaborators led by Professor Mark Achtman of the Warwick University School of Medicine and their pan-genomic badysis of ancient and modern Salmonella enterica demonstrates the genomic stability of the Invasive line para C for millennia. Current Biology .
He and his team badyzed the bacterial DNA found in the teeth and bones of the skeleton. of a young woman who had migrated to Trondheim from the northernmost regions of Scandinavia or northwestern Russia in her early teens only to die there at the age of 19-24.
They reconstructed a genome of Salmonella Paratyphi C that causes enteric fever in areas of poor hygiene and lack of clean drinking water. Their discovery indicates that the young Norwegian died of this disease and suggests that these bacteria have long caused enteric fever in northern Europe,
Prof Achtman said: " Paratyphi C is very rare today in Europe and North America except for occasional travelers from South and East Asia or from Africa, where the disease is more common C & # 39; is the first time salmonella has been found in ancient human remains in Europe, which is surprising because other salmonella are more common today, especially salmonella causing typhoid fever, called typhi, and salmonella causing food poisoning Earlier this year, Vågene and his coauthors described Paratyphi C Skeletons in Mexico, who died in 1545 CE, and speculated on Paratyphi C entered the Americas with Europeans, 19659003] The new findings included comparative genome badyzes of Paratyphi C found in the skeleton against the modern sequences of the Salmonella genome of EnteroBase, an online database developed at the University of Warwick and used internationally. This revealed that Paratyphi C represents the evolutionary descendants of a common ancestor, or clade, within the Para C lineage. The Para C lineage includes choleraesuis, which causes septicemia in pigs and wild boars, and Typhisuis that causes epidemic porcine salmonellosis (chronic paratyphoid) in domestic pigs. These different host specificities have probably evolved in Europe over the past 4,000 years and coincide with the time of domestication of pigs in Europe.
According to historical records, humans have long been affected by bacterial infections, but genomic badyzes of living bacterial pathogens commonly predict a date for the most recent common ancestor dating from a few centuries. In general, evolutionary trees contain a group of stems, which may include lines that are now rare or extinct, as well as the crown group of living organisms. Historical reconstructions based solely on the crown group ignore the older sublines in the stem group and thus provide an incomplete picture of the evolutionary older history of the pathogen. In contrast, ancient DNA badyzes like the genome of Paratyphi C can shed light on millennia of evolution of bacterial pathogens anterior to the origin of the crown group .
Professor Achtman added: "Using EnteroBase we have been able to define the Para C lineage from 50,000 modern genotypes of Salmonella enterica and discover that over its 3,000-year history, only a few genomic changes have occurred in the Para lineage. C.
"In addition to restating our understanding of Salmonella enterica, the research has sparked intriguing speculations about the historical hops of the host during the Neolithic between humans and their domestic animals.
UNIVERSITY OF WARWICK
Header Image – This is the reconstruction of the young woman's face Credit: Caroline Wilkinson, Mark Roughley, Ching Liu and Kathryn Smith at the Face Lab of Liverpool John Moores University for their different contributions to the production of the facial representation
 |
 |
[ad_2]
Source link