[ad_1]
A group of Harvard researchers recently extended the use of electronic cryomicroscopy (cryo-EM) to analyze small proteins at high resolution. In their study, published on the preprint server bioRxiv *, the researchers describe the method that enables the high-resolution structure determination of small proteins by single-particle cryo-EM using a set of target-binding nanobodies called Legobody.
 Study: Determination of the cryo-EM structure of small proteins by nanobody binding scaffolds (Legobodies). Image Credit: Dr Thomas Spaeter / Shutterstock.com
Study: Determination of the cryo-EM structure of small proteins by nanobody binding scaffolds (Legobodies). Image Credit: Dr Thomas Spaeter / Shutterstock.com
Strengths and limitations of cryo-EM
Single-particle electronic cryo-EM is an ideal technique for determining the structure of small proteins. Despite its usefulness, cryo-EM has been limited in its ability to analyze particles smaller than 100 kilodaltons (kDa). This poses a challenge when analyzing proteins less than 100 kDa, because about 50% of membrane proteins and other medically important proteins are less than 50 kDa.
Among the different approaches that have been used to image small proteins using cryo-EM, one method involves the fusion of the target protein with different scaffolds or the use of a binding partner to increase the size of the protein. Some of the different scaffolds that have been used for this purpose include modified ankyrin repeat proteins (DARPins), antibody Fab fragments or nanobodies.
What are nanobodies?
Nanobodies, which are derived from single chain antibodies from camels, are an attractive option for the fusion approach. Importantly, nanobodies can form rigid structures that bind to various forms of target proteins such as loops, convex surfaces, and cavities.
In addition, these scaffolds can be chosen from immune camelids or from large in vitro libraries displayed by phages, yeast cells or ribosomes.
Nanobodies can also be produced in large quantities quickly. Additional advantages of this approach include their ability to lock a protein in a fixed conformation and their existing extensive use in X-ray structural analysis.
Despite these advantages, the small size of the nanobodies, which is between 12 and 15 kDa, is a limiting factor for its direct use in cryo-EM. In an effort to overcome this problem, researchers in the present study increased the size of the nanobody to around 120 kDa by attaching two rigid scaffolds to this binding partner.
The usefulness of the Legobody
In their work, the researchers began by determining the structures of two small proteins, including the KDEL receptor (KDELR), which measures approximately 23 kDa. The second protein was the receptor binding domain (RBD) of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) spike protein, which is approximately 22 kDa. These two proteins are asymmetric monomers and are less than 40 kDa, which is well below the limit estimated for direct cryo-EM analysis of single particles.
KDELR is a small, unsymmetrical protein with no domains outside the membrane and a strong tendency to aggregate during sample preparation. In the present study, the researchers used additional “tricks” to reduce aggregation, improve stability, and ensure the preferred orientation of the particles. KDELR is representative of membrane proteins, many of which are of great interest in drug development.
Likewise, the RBD of the SARS-CoV-2 spike protein posed similar challenges for cryo-EM analysis, as it is composed of b strands and extended polypeptide segments. In addition to its small size, these characteristics make RBD a more difficult model compared to a protein consisting mainly of α-helices.
Here, the researchers obtained a good quality map, especially at the RBD / nanobody interface, with side chain density for all interacting amino acids.
Study results
The researchers showed an overall resolution of 3.2Å and 3.6Å with sufficient density to Again construction of models for the KDELR and SARS-CoV-2 RBD map. These resolutions confirmed the interactions between the two scaffolds and the nanobody.
In addition to the two scaffolds, both an Fab fragment of an antibody directed against the nanobody and a fragment of nanobody binding protein A fused to maltose binding protein (MBP). Fab binding domains have also been incorporated into Legobody. All interactions between these structures were rigid and maintained the distinct shape of the Legobody.
To this end, the Legobody consisted of two side arms formed by the two scaffolds, as well as a central lobe provided by the nanobody. Taken together, the Legobody had an overall size of around 120 kDa. In addition, the alignment center at the position of the nanobody facilitated all steps of cryo-EM analysis, from particle selection and classification to final refinement.
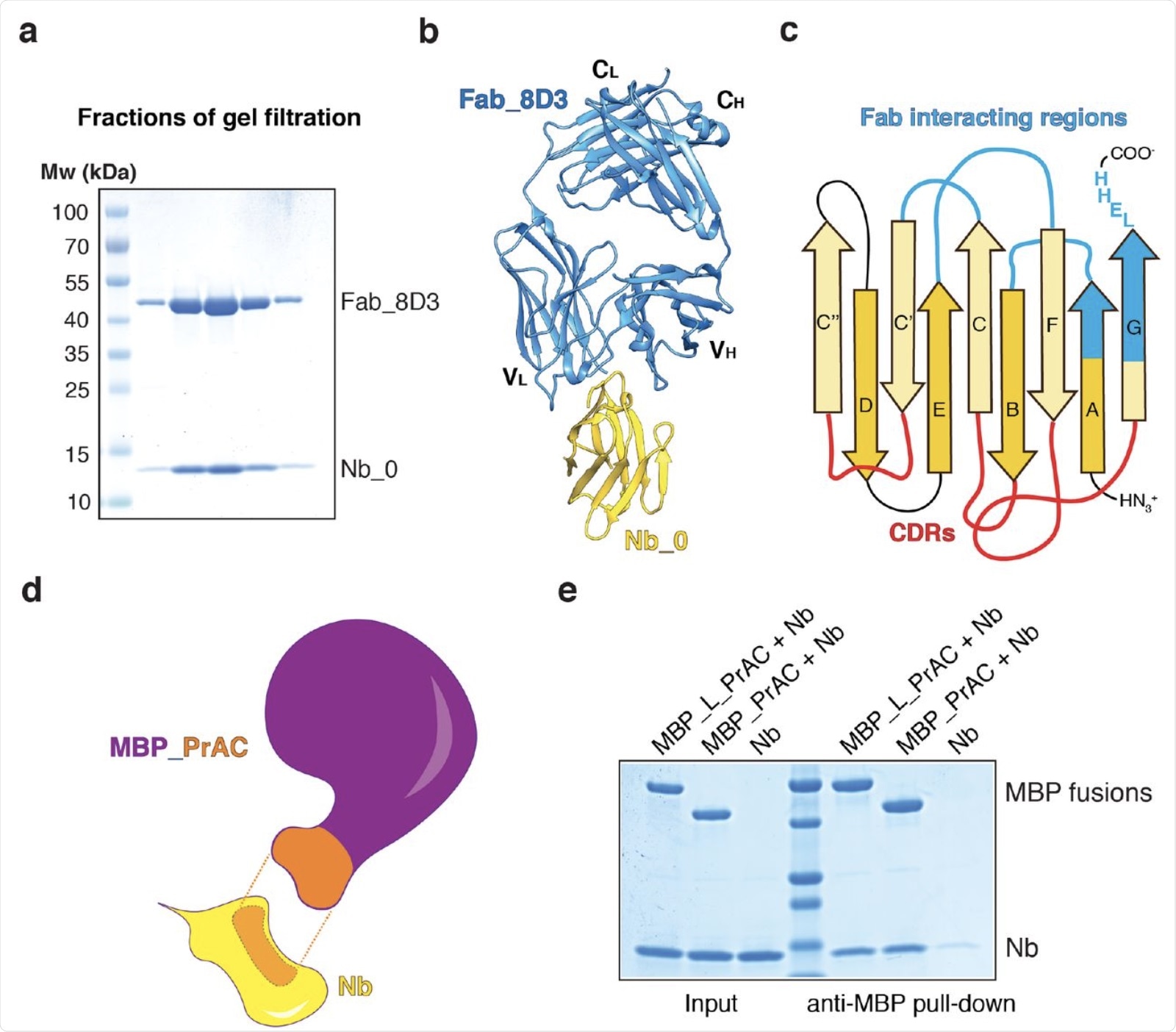 Generation of two scaffolds interacting with nanobodies. a. The Nb_0 nanobody was mixed with Fab_8D3 and the complex was subjected to gel filtration. Fractions were analyzed by SDS-PAGE and Coomassie blue staining. b, Crystal structure of the Nb_0 / Fab_8D3 complex shown in cartoon. Nb_0 is colored in yellow and Fab_8D3 in blue. c, Diagram of the nanobody, with strands marked according to the convention. The CDR loops are stained in red and the regions interacting with the Fab in blue. d, Diagram of the interaction of the nanobody (Nb; yellow) with the C domain of protein A (PrAC; orange) grafted onto the maltose binding protein (MBP; violet). The regions of interaction between Nb and MBP_PrAC are shown in orange with dotted lines in between. e, PrAC merged with MBP via a flexible linker (MBP_L_PrAC) or grafted on MBP (MBP_PrAC) was mixed with Nb. The mixture was incubated with amylose resin interacting with MBP, and the bound material was analyzed by SDS-PAGE and Coomassie blue staining. The entry corresponds to 50% of the material used for the pull-down.
Generation of two scaffolds interacting with nanobodies. a. The Nb_0 nanobody was mixed with Fab_8D3 and the complex was subjected to gel filtration. Fractions were analyzed by SDS-PAGE and Coomassie blue staining. b, Crystal structure of the Nb_0 / Fab_8D3 complex shown in cartoon. Nb_0 is colored in yellow and Fab_8D3 in blue. c, Diagram of the nanobody, with strands marked according to the convention. The CDR loops are stained in red and the regions interacting with the Fab in blue. d, Diagram of the interaction of the nanobody (Nb; yellow) with the C domain of protein A (PrAC; orange) grafted onto the maltose binding protein (MBP; violet). The regions of interaction between Nb and MBP_PrAC are shown in orange with dotted lines in between. e, PrAC merged with MBP via a flexible linker (MBP_L_PrAC) or grafted on MBP (MBP_PrAC) was mixed with Nb. The mixture was incubated with amylose resin interacting with MBP, and the bound material was analyzed by SDS-PAGE and Coomassie blue staining. The entry corresponds to 50% of the material used for the pull-down.
Conclusion
Overall, the Legobody approach discussed here is a sufficient method that allows the use of cryo-EM for the structural determination of small proteins. The researchers indicate that this new method could be used for any target binding after its tight binding to an available nanobody.
In particular, several limitations are associated with the current method. Additionally, it may be important to modify the Legobody in the future to increase its molecular weight, stability, and stiffness in order to ultimately improve the resolution of cryo-EM maps.
*Important Notice
bioRxiv publishes preliminary scientific reports which are not peer reviewed and, therefore, should not be considered conclusive, guide clinical practice / health-related behavior, or treated as established information.
Source link