[ad_1]
Australian scientists recently deciphered the structural and molecular basis for T cell recognition of a dominant epitope on the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein. The results reveal that the dominant epitope derived from the peak induces a polyfunctional response of CD8 + T lymphocytes in individuals recovered from coronavirus disease 2019 (COVID-19). The study is currently available on the bioRxiv* preprint server.
During viral infection, cytotoxic CD8 + T cells play an essential role in the recognition and elimination of the invading virus. Studies have shown that CD8 + T cells are able to recognize and effectively eliminate SARS-CoV-2 variants in COVID-19 patients.
Mechanically, CD8 + T cells recognize viral epitopes presented by human leukocyte antigen (HLA) class I molecules through specific T cell receptors (TCRs), leading to the initiation of a cascade of immune reactions which ultimately result in viral clearance. The most common HLA class I molecule in the world population is HLA-A * 02:01. A growing body of evidence has indicated that individuals recovered by COVID-19 with HLA-A * 2:01 demonstrate a robust CD8 + T cell response to an immunodominant SARS-CoV-2-derived epitope, namely YLQ.
The study
Scientists have described the X-ray crystallographic structure of a public complex of TCR – YLQ epitopes. In addition, they characterized the response of CD8 + T lymphocytes to the YLQ epitope presented by HLA-A * 02:01 in individuals recovered by COVID-19. A public TCR is an epitope-specific TCR shared between unrelated individuals.
Response of CD8 + T cells to the YLQ epitope
Scientists determined the immunogenicity of the YLQ epitope in three individuals recovered from COVID-19. By growing CD8 + T cells against the YLQ epitope, they observed variable YLQ-specific T cell response and cytokine production between participants. More specifically, only two out of three participants had CD8 + T cells capable of producing all four cytokines (IFN, TNF, CD107a and MIP1β); however, only two cytokine-producing CD8 + T cells were detected in the other participant.
Taken together, these observations indicate that after recovery from COVID-19, individuals are able to induce a polyfunctional CD8 + T cell response to the YLQ epitope; however, the level of polyfunctionality may differ from individual to individual.

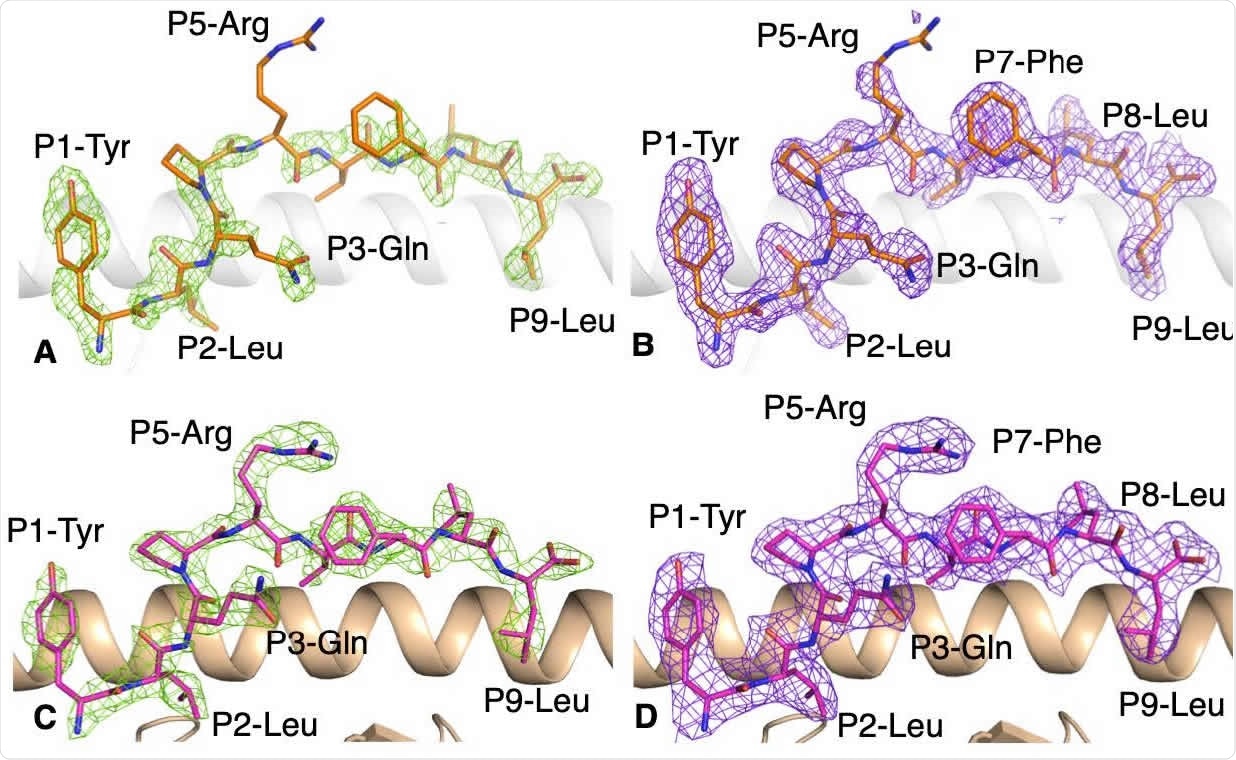
Electron density map for the YLQ peptide bound to HLA-A * 02:01 without and with the YLQ-SG3 TCR. 219 (A, B) Electronic density map of (A) Fo-Fc map at 3σ (green) and (B) 2Fo-Fc at 1 (purple) around peptide YLQ 220 (orange stick) in complex with HLA-A * 02:01 (white drawing). (C, D) Electron density maps of (C) Fo-Fc map at 3σ 221 (green) and (D) 2Fo-Fc at 1 (purple) around the YLQ peptide (pink stick) presented by HLA-A * 02:01 (beige 222 cartoon) bound for the YLQ-SG3 TCR.
Presentation of the YLQ epitope by HLA-A * 02: 01
Scientists aligned spike sequences from SARS-CoV-2 and seasonal coronaviruses to determine whether the YLQ epitope is conserved between human coronaviruses. They found that the epitope shares only four residues with seasonal coronaviruses. However, they observed that the epitope is highly conserved between the different variants of SARS-CoV-2 with less than 0.5% mutations of its residues.
By analyzing the X-ray crystallographic structure of the HLA-A * 02: 01 – YLQ complex, they revealed that the epitope maintains a stable and rigid conformation in the HLA slit and that the amino acid residues of the epitope have long side chains that potentially interact with TCRs.
Recognition of the YLQ epitope by T cell receptors
From previously available studies, the scientists analyzed the TCR sequences of YLQ-specific clonotypes observed in individuals recovered from COVID-19. Their analysis revealed a strongly biased TCR repertoire among unrelated HLA-A * 02:01 positive individuals. Additionally, they noticed a public representation of TCR among unrelated individuals and selected it to further characterize the T cell response to the YLQ epitope.
By analyzing the structure of the YLQ – public TCR complex, they noticed that the receptor has a high binding affinity for the HLA-A * 02:01 – YLQ complex. During interaction, the public TCR docks diagonally above the center of the YLQ epitope. The TCR alpha chain contributes about 67% of the interaction. It is important to note that minimal structural rearrangements occur when docking the public TCR to the HLA-A * 02:01 – YLQ complex.
Importance of the study
The study describes the structural and molecular basis of the interaction between a public TCR and an immunodominant YLQ epitope derived from SARS-CoV-2. In individuals recovered from COVID-19, YLQ-specific T cells exhibit a bias in their TCR repertoire because the TCR alpha chain contributes 67% of the interaction, the CDR1 / 2a loops (regions determining complementarity responsible for the specificity of the TCR) contributing to 40% of the total interactions.
As the scientists mentioned, the YLQ epitope can be used as a potential biomarker for SARS-CoV-2 infection because the epitope is highly immunogenic in most individuals recovered from COVID-19 and unrecognized in healthy individuals. Additionally, the ability to stimulate a strong polyfunctional CD8 + T cell response makes the YLQ epitope a promising target for T cell therapies or COVID-19 vaccines.
Source link