[ad_1]
Various strains of coronavirus (CoV) can infect both animals and humans and cause different diseases. CoVs are known to have caused three epidemics and pandemics in the past two decades. Therefore, knowledge about the evolution and emergence of CoV diversity in animals and humans is important.
 Study: Evolutionary dynamics and epidemiology of endemic and emerging coronaviruses in humans, pets and wildlife. Image Credit: joshimerbin / Shutterstock
Study: Evolutionary dynamics and epidemiology of endemic and emerging coronaviruses in humans, pets and wildlife. Image Credit: joshimerbin / Shutterstock
Coronavirus disease 19 (COVID-19) was first identified in China in December 2019, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Later in March 2020, it was declared a global pandemic by the World Health Organization (WHO).
The four genera of coronavirus include alpha-coronavirus (α-CoV), beta-coronavirus (β-CoV), gamma-coronavirus (γ-CoV), and delta coronavirus (δ-CoV). All four subtypes belong to the Orthocoronaviridae. Α- and -CoV are known to infect mammals, γ-CoV infects avian species while δ-CoV infects both mammalian and avian species.
A study published in the Virus journal aimed to understand the evolutionary and epidemiological dynamics of coronavirus diversity in animals, humans and wildlife at ecosystem interfaces.
The study
The study involved a detailed literature search using specific keywords in PubMed, Scopus, Google Scholar and Web of Science on the natural infection of coronaviruses in humans, animals and wildlife. Gray literature was also searched for the diversity of human and animal coronaviruses. The literature was selected based on their natural infection from different species. In addition, information regarding the number of cases and the chronology of emergence of different animal and human CoVs was also collected.
Several phylogenetic trees have been prepared to determine ancestral relationships between human and animal coronaviruses. Several representative CoV sequences from humans, animals and wild animals were aligned and finally evaluated by phylogenetic analysis.
Study results
Emerging and endemic CoV infections in livestock and pets
A wide range of farmed CoVs produce different diseases in different domestic animals, but not all of them have zoonotic potential. Bovine coronavirus (BCoV) has a wide host range and is also genetically related to human β-CoV OC43. In addition to BCoV, a critical porcine coronavirus, porcine acute diarrhea syndrome virus (SADS-CoV) can pose a serious threat to human health. However, the nucleotide similarity between porcine CoV and SARS-CoV-2 is low, suggesting that SARS-CoV-2 did not emerge from pigs.
There is a strong similarity between porcine angiotensin converting enzyme 2 (ACE2) and human ACE2, suggesting that SARS-CoV-2 is capable of infecting pigs. Although pigs are not susceptible to SARS-CoV-2, transmission and mutation between species can occur with porcine CoV.
Emerging CoVs in wildlife
Bats are known to be the ancestors of SARS, MERS, SARS-CoV-2 and other important CoVs that threaten human and animal health. Palm civets and raccoon dogs have been shown to be intermediate hosts for SARS-CoV-like viruses. Additionally, civet strains of SARS-like CoV were closely related to the human strains that led to the 2002-2003 SARS epidemic.
The results also indicated that several wild-species infections of SARS-CoV-2 originated in humans. These wild animal species may contribute to the future evolution and transmission of CoV to other susceptible animals. For example, infection in mink was initiated by humans, which led to the emergence of a new variant of mink. This leads to the persistence of the virus in nature and its emergence from time to time.
Marine CoVs have been shown to be very similar to human coronaviruses. As a result of human infection, there is a high probability that SARS-CoV-2 will be released into the marine environment. SARS-CoV-2 nucleic acid has been detected in sewage and wastewater to be transmitted to wild aquatic animals.
Emerging and endemic CoV infection in humans
Endemic human CoVs include OC43, 229E, NL63, and HKU1. All are of animal origin whose spillover effects have already been proven. Three pandemic CoVs, SARS, MERS and SARS-CoV-2, originated from animals, primarily Rhinolophus bats.
The ongoing pandemic, caused by SARS-CoV-2, has spread from a seafood and wildlife market in China. Initially, the infection occurred from contact with animals or the environment, resulting in animal-to-human transmission, but recent studies have shown that human-to-human transmission occurs through respiratory droplets. Nosocomial infections are also a major cause of infection with SARS, MERS and SARS-CoV-2.
Structural analysis of SARS-CoV-2 has shown that although they use the same receptor for input as SARS-CoV, the affinity for the receptor is much higher in the case of SARS-CoV-2 . This indicates that SARS-CoV-2 could be more infectious to humans than SARS-CoV.
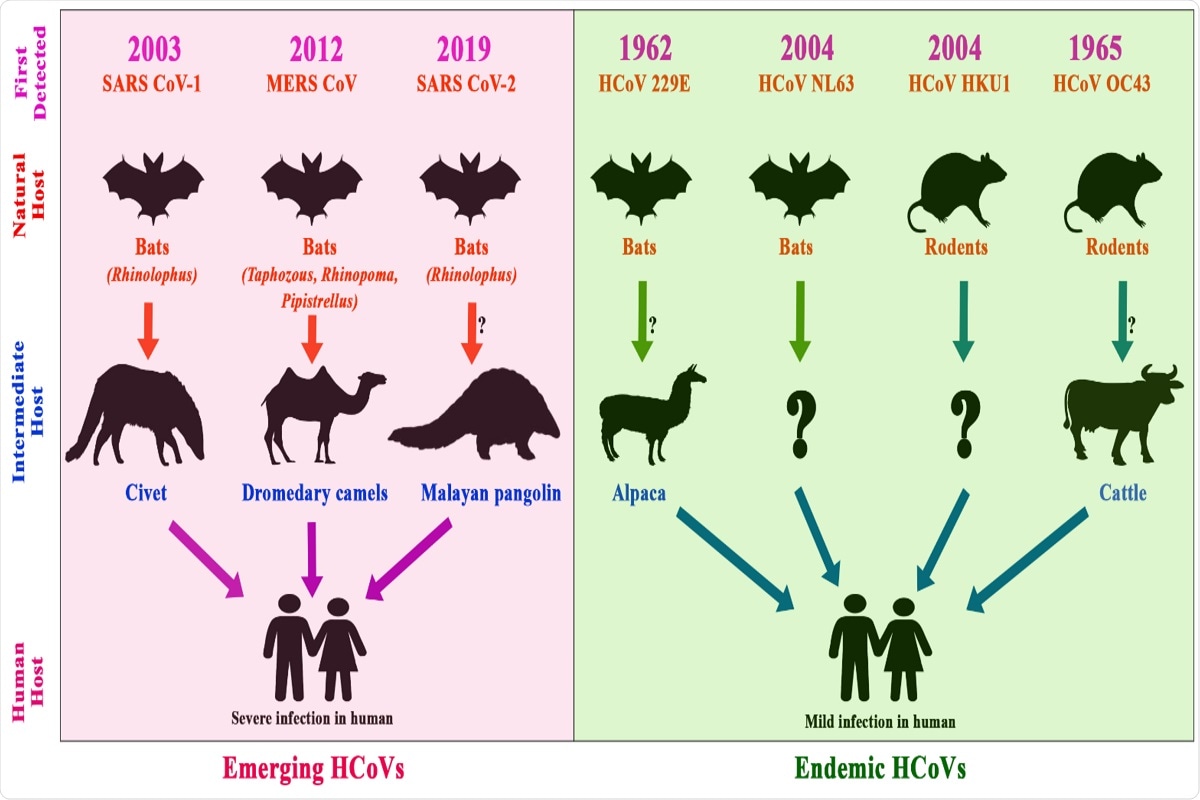 Figure 5. Chronology of the emergence of human CoVs, their reservoirs and their intermediate hosts; the pink shaded area represents emerging coronaviruses, while the green shaded area represents endemic coronaviruses. The arrows show the route of virus transmission from animals to humans via intermediate hosts.
Figure 5. Chronology of the emergence of human CoVs, their reservoirs and their intermediate hosts; the pink shaded area represents emerging coronaviruses, while the green shaded area represents endemic coronaviruses. The arrows show the route of virus transmission from animals to humans via intermediate hosts.
Conclusion
The present study has shown that many animals, both wild, domestic and human, are susceptible to various CoVs. There is considerable evidence of inter-species transmission which leads to new strains and increased transmissibility of CoVs. The strains of coronavirus that cause pandemics and endemics have had an impact on public health, the global economy and society. Strengthening animal surveillance could prevent the appearance of CoVs of animal origin.
Journal reference:
- Islam, A. et al. (2021) “Evolutionary dynamics and epidemiology of endemic and emerging coronaviruses in humans, domestic animals and wildlife”, Virus, 13 (10), p. 1908. do I: 10.3390 / v13101908.
Source link