
[ad_1]
The antibiotic resistance of bacteria is spreading rapidly around the world and has even been described as a global threat to humans as serious as climate change. Excessive and imprudent use of antibiotics is usually implicated, but the rate and extent of spread can not be explained by excessive consumption.
Antibiotic resistance has now been found in isolated parts of the world where humans and antibiotics are rare or absent. We recently published a study in Environment International reporting antibiotic-resistant genes in Svalbard in the High Arctic.
Antibiotic resistance itself is natural, but continued human exposure to antibiotics has selected stronger and stronger bacteria. These bacteria often acquire antibiotic resistance genes that allow them to make powerful defense proteins. As antibiotics are used, new forms of resistance are evolving, including multiple resistance to pathogens, creating a health crisis.
David Graham, Author provided
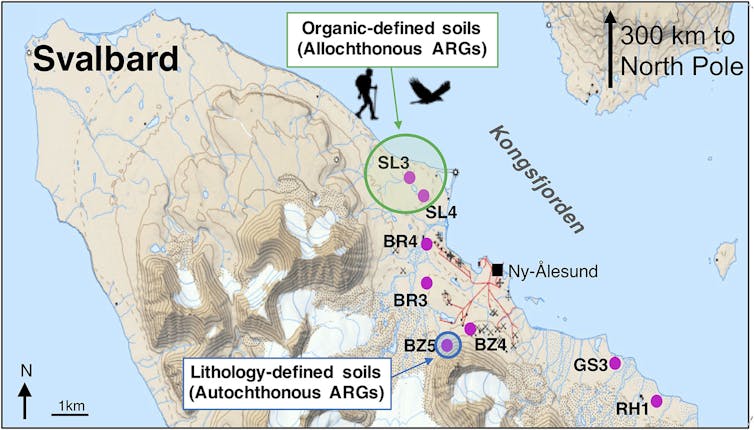
For our research, we extracted bacterial DNA from 40 soil cores located at eight locations along Kongsfjorden on the west coast of Spitsbergen, in the Arctic Ocean, 300 km from the North Pole. This place is quite unusual in the Arctic for its vibrant wildlife populations and nearby fjord that does not freeze. We examined Arctic soil DNA and quantified 131 antibiotic resistance genes badociated with nine major clbades of antibiotics. These include aminoglycosides, carbapenem, macrolides and β-lactams, many of which are used to treat human and veterinary infections.
Some of the genes detected, some of which are almost certainly not "local" in the Arctic, can confer resistance to several drugs. These "foreign" genes have been found in the highest concentrations near freshwater sources – areas where wildlife tends to congregate.
An antibiotic resistance gene that we found is called "blaNDM-1". This gene gives bacteria resistance to carbapenem antibiotics, one of our last recourse against infectious diseases. We found this gene in soils near a small Arctic lake in 2013, but it was first detected in a patient hospitalized in India in 2007 and then found in surface water in urban India in 2010 .
Although not explicitly Indian, BlaNDM-1 was first identified from one side of the world and migrated from the other side in less than three years to end up in a place where few humans reside and where antibiotics are rare. How did it happen?

David Graham, Author provided
Migrating resistance
Places where sanitation is inadequate often have higher levels of resistance genes and bacteria in the surrounding water and soil. Local overcrowding, the use of poorly controlled antibiotics and incomplete treatment of sewage make it easier to choose resistance in these places.
Local water and food resources are contaminated with feces, allowing resistance genes and their bacterial hosts to be ingested by humans and wildlife – from an excursion to the sea. 39; other from one region of the world to the other. It is not known whether it is a single migration by a bird or a human, or a chain reaction of exhibitions. Whichever path is chosen, resistance genes move rapidly and in places where antibiotics are not present.
When antibiotic resistance migrates, it also changes as it pbades through animals and the environment in general. This complicates the search for these genes from their origins to their final destination. In the case of blaNDM-1, it was initially detected as a single gene in a few places, but many variants have been found in dozens of countries, including the Arctic.
If resistance genes migrate through humans or wildlife to other places, especially in places where local sanitation is inadequate, such genes and bacterial hosts could be selected from among the subsequent human and wildlife populations. That's what we think is happening around the world. These genes move around the world with humans and other animals, sowing new places with potential for resistance.
Nokwalai / Shutterstock
Reduce the use of antibiotics is essential to fight bacterial resistance. In the UK, this is described in the five-year action plan announced by Health Secretary Matt Hanbad. However, reducing the use of local antibiotics may have a limited effect on overall resistance unless environmental pathways are not further considered. Improving sanitation and water quality around the world must be part of the fight against antibiotic resistance.
However, our study on the High Arctic has a positive rating. Although we detected genes such as blaNDM-1, we also found other interesting and unexpected genes for antibiotic resistance. We found a gene coding for multiple drug resistance in tuberculosis bacteria in almost all of our soil nuclei. This gene was not explicitly related to fecal matter, which means that it is almost certainly natural for this remote environment.
Read more:
It's the era of the antibiotic revolution, not the apocalypse
This may sound like bad news, but it tells us where this specific resistance gene could come from, and better knowledge of this gene could help us fight resistant TB in the future. It also suggests that information from distant soils such as Svalbard could be a valuable source of undiscovered drugs.
That said, focusing efforts on developing new arsenals of antibiotics may not be enough. It would also be wise for the richest countries to help the poorest to improve the quality of water and sanitation, even if it only provides toilets to reduce defecation at the same time. ;outdoors. Smarter use of antibiotics in agriculture and medicine is a necessary step to combat resistance, but only a comprehensive approach will reduce the evolution and spread of antibiotic resistance.
Source link