
[ad_1]
One day in the future, you will be able to take a pill to cure a disease – and you owe your recovery to the tiny microbes that bloom in the slippery layer of mucus that covers the fish.
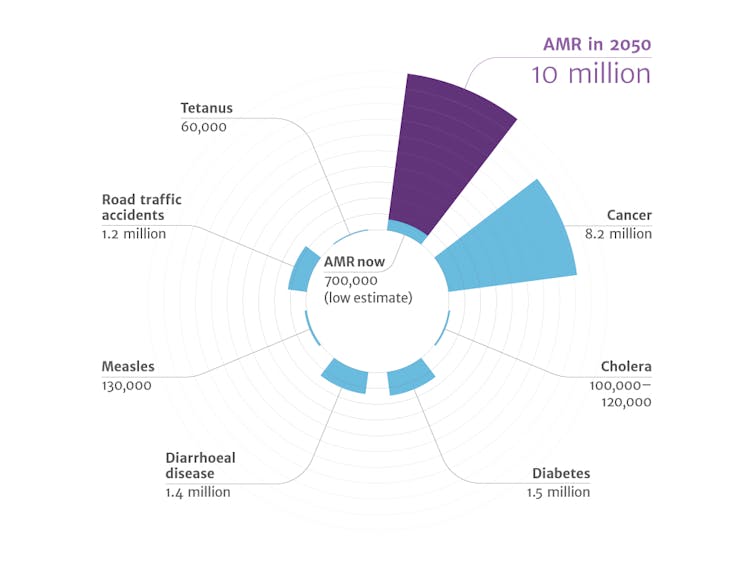
It is essential to find the next generation of antibiotics. The incidence of bacterial infections resistant to current antibiotics continues to increase. The World Health Organization has warned that this problem would only worsen, and a recent study predicts that by 2050, drug-resistant infections will affect more people than cancer.
Review of antimicrobial resistance, CC BY
But how do you find a new antibiotic?
It is perhaps surprising that more than 70% of the currently used anti-infectives are derived from natural chemicals. Plants and microbes produce a wide range of complex chemicals, some of which have antibiotic or antiviral properties, or even are toxic to cells. For example, amoxicillin is one of the most commonly prescribed antibiotics and is a derivative of a chemical isolated from Penicillium mold.
Although many efforts to identify new anti-infectives have focused on soil microbes, these surround us. In fact, they are everywhere on us and inside us. Animals, including humans, harbor a diverse community of microbes on the skin and in the gastrointestinal system.
There is a growing consensus that these microbes can interact positively or negatively with host organisms, including facilitating digestion and reducing pathogenic infections, but also contributing to certain types of diseases. These microbes can also be a source of new antibiotics. For example, researchers have recently identified a new antibiotic from a bacterium found in the nose of humans.

FWC Fish and Wildlife Research Institute, CC BY-NC-ND
In my laboratory at Oregon State University, we have worked to identify the next generation of antibiotics from animal-related microbes. Our current efforts focus on the most diverse group of vertebrate, marine and freshwater fish. More than 33,000 species of fish have been identified, more than the sum of all other vertebrates on Earth. These animals often live in harsh environments and are susceptible to microbes that help them resist infection.
We are working with marine biologist Misty Paig-Tran of California State University Fullerton to obtain mucus samples from different Pacific fish species. On several trawls, her team was able to catch coastal fish and some deep-sea fish, totaling about 17 species. For example, they brought back several species of pink pearl from coastal waters and deeper deeper waters.

Loesgen Lab, CC BY-ND
The viscous mucus that covers the fish acts as a protective coating. When the animal moves in the water, it can come in contact with all kinds of bacteria, fungi, viruses and more. mucus acts as a physical barrier. The researchers believe that there is also a chemical component produced by the fish microbiome that helps prevent infection.
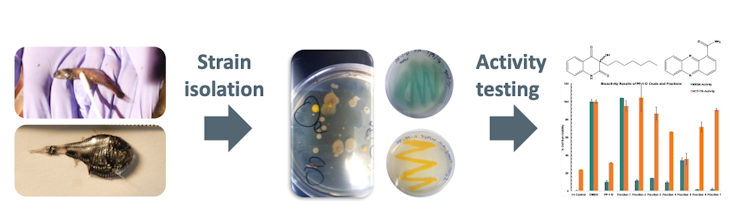
My collaborators and I were looking for interesting bacteria that we could isolate from the fish. Our goal was to explore the bioactivity in the bacterial extract in the hope of being able to exploit it for our own uses.

Loesgen Lab, CC BY-ND
We are currently studying the taxonomy of bacteria – that is, how are they related and how should they be clbadified in the tree of life? What species are they? Undergraduate researcher Molly Austin and chemistry graduate student Paige Mandelare were able to isolate 47 different bacterial strains from these fish mucus swabs. We cultivated them, extracted the chemicals they made, and then tested them to see if they inhibited common human pathogens.
Interestingly, we found that several bacterial extracts had high antimicrobial activity, with 15 extracts showing strong inhibition of methicillin resistance. Staphylococcus aureus. MRSA is a drug-resistant human pathogen that causes many infections that are difficult to treat in humans.
Loesgen Lab, CC BY-ND
We performed additional tests and badyzes on one of the most potent extracts and found that the microbes produced multiple badogs of a particular heterocyclic aromatic compound called phenazine, which had antibiotic activity. Motivated by these results, we checked whether the compounds of these extracts could also affect the cancer cells. We found that this fish derived Pseudomonas The bacterium, isolated from a coastal pink surfperch, also produced a metabolite that inhibited the growth of human colon carcinoma cells.
This research is ongoing in my lab and others, and the question of whether an active compound is an effective drug depends on many factors. However, these results suggest that fish-badociated microbes produce a wide range of diverse and complex chemicals and are an excellent source of effort for drug discovery.
Source link