[ad_1]
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which is the virus causing coronavirus disease 2019 (COVID-19), is a highly infectious positive strand ribonucleic acid (RNA) virus belonging to the family Coronaviridae. Among RNA viruses, coronaviruses contain the largest genome and are associated with complex replication processes.
The complexity of coronavirus replication is due to the fact that this process involves a frame shift slip, during which replicase jumps and multiple RNA duplexes are generated. Coronaviruses also develop numerous membrane compartments to protect their viral components, which increases the efficiency of replication and helps these viruses escape innate immune recognition.
 Study: Multiscale Correlative Cryoimaging Unveils Assembly and Release of SARS-CoV-2. Image Credit: CROCOOTHERIE / Shutterstock.com
Study: Multiscale Correlative Cryoimaging Unveils Assembly and Release of SARS-CoV-2. Image Credit: CROCOOTHERIE / Shutterstock.com
The structural proteins of SARS-CoV-2
SARS-CoV-2 is made up of four different structural proteins, which include spike (S), membrane (M), envelope (E), and core (N) proteins. The main function of the S protein, which is made up of the S1 and S2 domains, is to mediate the entry of the virus into the host cell.
SARS-CoV-2 infection involves two stages of pre-fusion and post-fusion. While pre-fusion involves both S1 and S2 subunits, the latter involves the S2 subunit alone.
The main role of the SARS-CoV-2 N protein is to encapsulate and protect the viral genome. Protein E is the smallest structural protein and acts as an ion channel. Finally, the transmembrane M protein, which is the structural protein most abundantly present in SARS-CoV-2, lines the inner surface of the lipid membrane of the virus.
SARS-CoV-2 replication cycle
The first stage of infection with SARS-CoV-2 involves the interaction between the S protein of the virus and the angiotensin-converting enzyme 2 (ACE2) receptor present on the surface of the host cell. Subsequently, S2 either cleaves at the cell surface in the presence of the cell protease TMPRSS2 or triggers endocytosis of the viral particles in the absence of TMPRSS2.
When the viral components are introduced into the cytoplasm of the host cell, the precursor polyproteins Pp1a and Pp1ab are synthesized. SARS-CoV-2 non-structural proteins such as nsp3, nsp4 and nsp6 rapidly induce the formation of membrane compartments called double membrane vesicles (DMVs). The processes of replication and transcription of the viral genome occur in these membrane compartments.
Scientists are curious about the mechanism by which messenger RNAs (mRNAs) reach the cytoplasm to be translated by the cellular ribosomes of sealed DMVs. A recent study on murine hepatitis coronavirus (MHV) and SARS-CoV-2 reported the presence of a molecular pore that served as an export portal for the transfer of mRNA and viral genome copies to positive strand.
Previous studies have also reported that the assembly of viral particles takes place at the level of modified cell membranes derived from the endoplasmic reticulum (ER), the Golgi apparatus and the ER-Golgi intermediate compartment (ERGIC). These studies further added that the virus is released by exocytosis.
Although extensive research has been conducted on the structural components of SARS-CoV-2, there is a limited amount of structural and ultrastructural evidence on the progression of SARS-CoV-2 infection in native cells. Additionally, there is a research gap related to understanding viral assembly and release.
A multi-scale cryo-correlative platform to visualize SARS-CoV-2 infection
A new study published in Nature Communication correlated cytopathic episodes triggered by SARS-CoV-2 with virus replication processes in frozen hydrated cells. The authors of this study used a unique correlative, multimodal, multiscale cryoimaging technique to examine the progression of SARS-CoV-2 replication using Vero cells.
The technique used in the present study is unique in that it provides a holistic picture of SARS-CoV-2 infection that includes the entire cell fused to individual virus spike molecules. This approach also revealed pathways associated with virus assembly, as well as the egress and cytopathic effects of SARS-CoV-2 infection.
In this study, the researchers combined different techniques including cryogenic serial focused ion beam scanning electron microscopy with scanning electron microscopy (cryoFIB / SEM) and soft X-ray tomography to study the morphology of the body. whole cell. The data obtained from these techniques were then correlated with high resolution cryoelectronic tomography (cryoET). One of the advantages of the cryoET is the use of frozen-hydrated conditions which provide quick and easy sample preparation.
The images obtained on the replication of SARS-CoV-2 revealed a spatially well organized and very efficient process. Scientists observed that every step of the replication process, including genome replication, protein synthesis, and virus assembly, to name a few, takes place in specific cytoplasmic compartments.
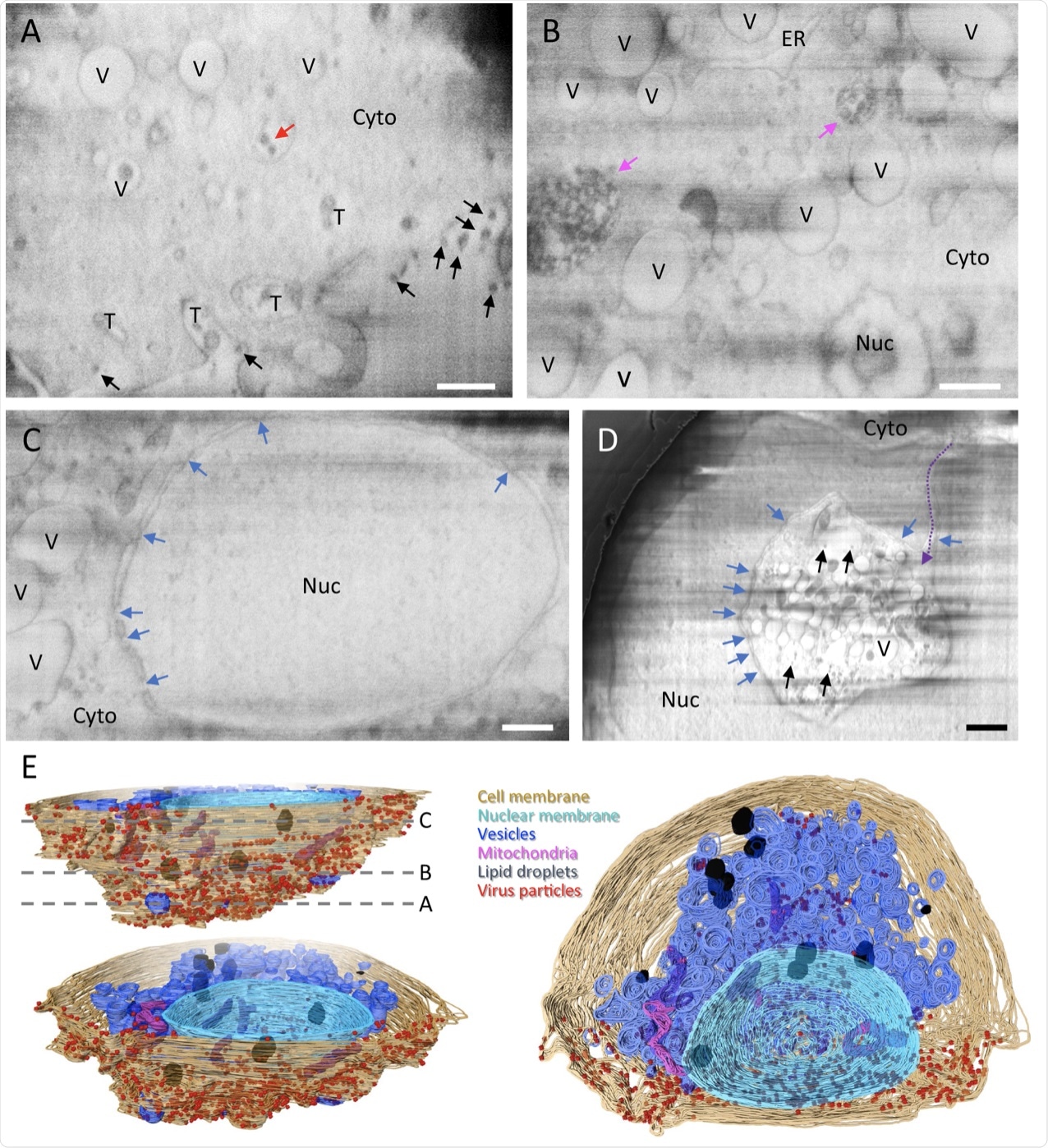 A–D Representative cryoFIB / SEM slices of a SARS-CoV-2 infected cell at the periphery of the cell (A), cytoplasm (B), cell nucleus (VS), and invagination of the cytoplasm into nuclear space (note, from a different cell) (D). Scale bars, 500 nm in A–VS, 1 µm in D. Black and red arrows, extracellular and intracellular viral particles, respectively; blue arrows, nuclear pores; pink arrows, complex membrane compartment; purple dotted arrow, invagination path. Vesicles V, T tunnels, Nuc nucleus, cytoplasm, endoplasmic reticulum of the ER. E Surface rendering of the segmented volume of the SARS-CoV-2 infected cell shown in A–VS. Segmented organelles and viral particles are labeled with the colors shown. The dotted lines (E, upper left panel) indicate the positions of the slices displayed in A–VS, respectively.
A–D Representative cryoFIB / SEM slices of a SARS-CoV-2 infected cell at the periphery of the cell (A), cytoplasm (B), cell nucleus (VS), and invagination of the cytoplasm into nuclear space (note, from a different cell) (D). Scale bars, 500 nm in A–VS, 1 µm in D. Black and red arrows, extracellular and intracellular viral particles, respectively; blue arrows, nuclear pores; pink arrows, complex membrane compartment; purple dotted arrow, invagination path. Vesicles V, T tunnels, Nuc nucleus, cytoplasm, endoplasmic reticulum of the ER. E Surface rendering of the segmented volume of the SARS-CoV-2 infected cell shown in A–VS. Segmented organelles and viral particles are labeled with the colors shown. The dotted lines (E, upper left panel) indicate the positions of the slices displayed in A–VS, respectively.
A multi-scale view of SARS-CoV-2 genome replication
The researchers in this study illustrated five different steps associated with the replication of the SARS-CoV-2 genome, which include:
- Step 1: RNA replication occurs in DMVs, thereby separating vRNA and subgenomic mRNA from the host cell’s innate immune response.
- 2nd step: Newly synthesized vRNAs are transported from DMVs to virus assembly sites through transmembrane portals. MRNAs are also transported through the same portals to the cytoplasm, where it produces proteins.
- Step 3: Viral S proteins are transported to assembly sites by small transport vesicles.
- Step 4: These vesicles fuse with single membrane vesicles (SMVs), where viral spikes come together at the assembly site and the budding process begins. These events occur in the presence of vRNA. N proteins are also formed in this step.
- Step 5: After the assembly and budding processes, a vesicle containing the virus is formed which may contain several virus particles. These particles can then exit through tunnels or through lysosomal exocytosis.
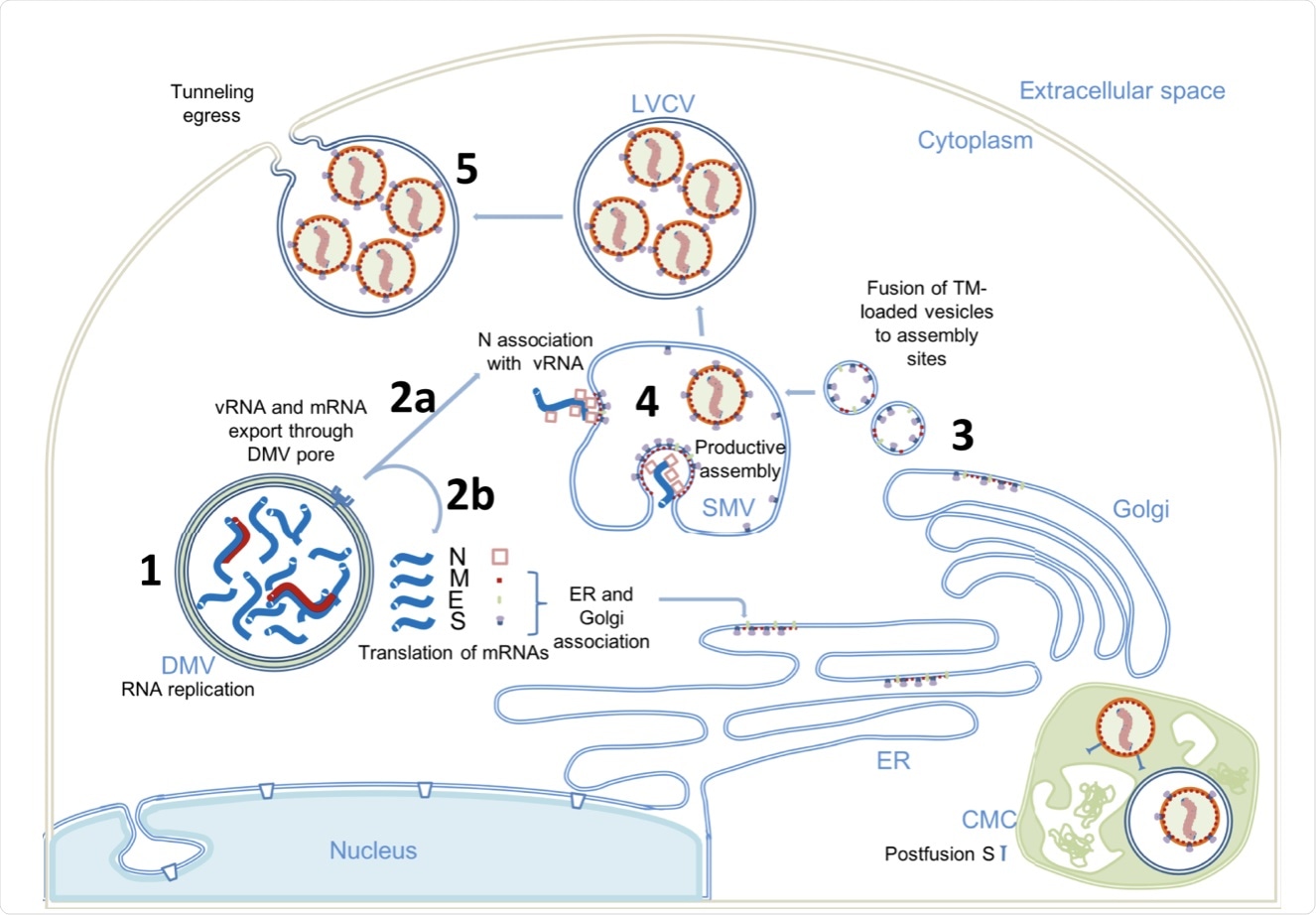 (1) Replication of the viral genome occurs within DMVs, generating negative stranded viral and subgenomic RNAs (red), positive genomic copy of vRNA, and subgenomic mRNAs (blue). (2) Positive RNAs are exported to the cytoplasm through the pores of the DMV. The subgenomic mRNAs are translated (2b). The structural proteins M, E and S associate with the ER and Golgi membranes. The genomic vRNA becomes complexed with the newly synthesized N (2a). (3) S, E and M are transported in dense vesicles, which are fused with VMS. (4) Productive viral assembly occurs in the VMS, aggregating viral tips and packaging the genome into RNPs. Viruses bud in the internal space of the VMS. (5) The exit is via tunnels. The complex membrane compartment (CMC) is shown in green, which contains both viral particles containing postfusion spikes and viral particles containing prefusion spikes. DMV double membrane vesicle, SMV single membrane vesicle, CMC complex membrane compartment, large vesicle containing LVCV virus.
(1) Replication of the viral genome occurs within DMVs, generating negative stranded viral and subgenomic RNAs (red), positive genomic copy of vRNA, and subgenomic mRNAs (blue). (2) Positive RNAs are exported to the cytoplasm through the pores of the DMV. The subgenomic mRNAs are translated (2b). The structural proteins M, E and S associate with the ER and Golgi membranes. The genomic vRNA becomes complexed with the newly synthesized N (2a). (3) S, E and M are transported in dense vesicles, which are fused with VMS. (4) Productive viral assembly occurs in the VMS, aggregating viral tips and packaging the genome into RNPs. Viruses bud in the internal space of the VMS. (5) The exit is via tunnels. The complex membrane compartment (CMC) is shown in green, which contains both viral particles containing postfusion spikes and viral particles containing prefusion spikes. DMV double membrane vesicle, SMV single membrane vesicle, CMC complex membrane compartment, large vesicle containing LVCV virus.
Source link