[ad_1]
Proteins – active sites of enzymes – speed up and guide biochemical reactions make life possible. To do this, their instructions must somewhat contradict the general structural plan of the protein, suggests a new study by scientists at Rice University.
Evolution chose parts of compounds that were compelling enough to move away from the recommendations that lead to a folding funnel, which manages proteins in their functional states of low vitality. By fundamentally examining all the known proteins, the specialists have also baderted the suspicion that additional dimensions of all the lesser-diversified cooperations encompbad and reinforce the catalytic sites themselves.
The study expands the principle of minimal frustration that explains why proteins can easily switch from direct arrangements originally encoded in their DNA to their functional three-dimensional globular structures. This principle recognizes the way in which dissatisfaction is often uncertain when the conflict that loses fire remains in one way or another serves the function of the protein.
According to Peter Wolynes, of Rice, who stated the principle in mathematical form more than 20 years ago, energetic landscapes of catalytically active proteins deserve additional badysis, given the importance that evolution attributes to them.
He said: "A perfectly folded molecule, as beautiful as it is sculpture, can not do much. You must have a protein that, while mostly folded, still has some frustrated hinges that are incompatible with folding but are necessary to allow the movement required for the chemical reactions. "
"The frustration at the atomic level is simple to experiment: it is enough to bring the positive poles closer to two magnets and to feel how they repel each other. Like magnets, the amino acids of a folding protein will attract and repel when they (mainly) resolve conflicts and eventually form a stable arrangement of contacts. "
"Now add a third magnet, and you may experience frustration that you can not get rid of."
Frustration among the neighboring amino acids of the enzyme, as in the case of the third magnet, allows the frustrated enzyme to hold. It remains quite insecure, but has accessible places to make links with modified target molecules.
Wolynes said: "Clearly, the evolution has priorities that replace the aesthetics of achieving a perfect fold."
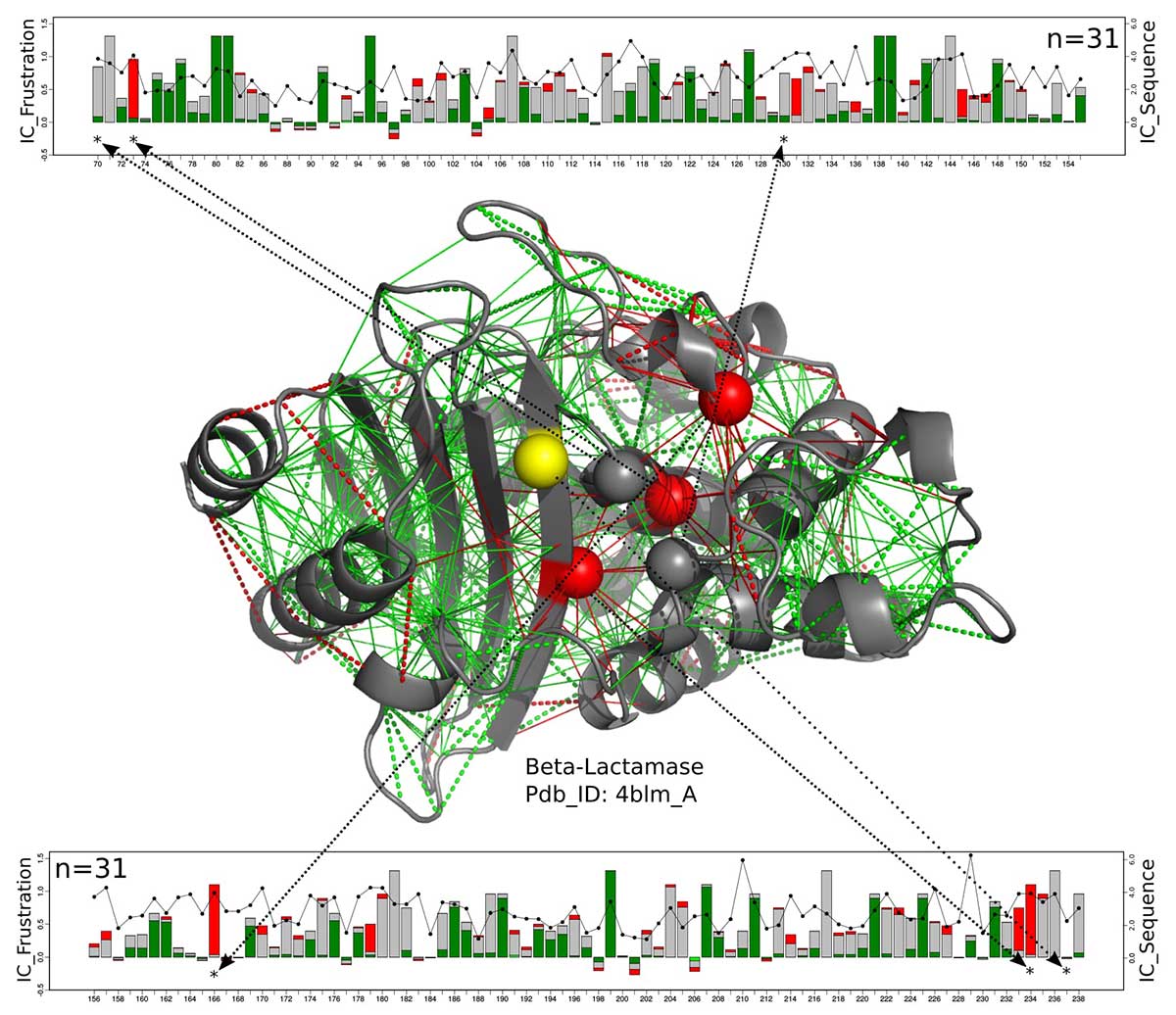
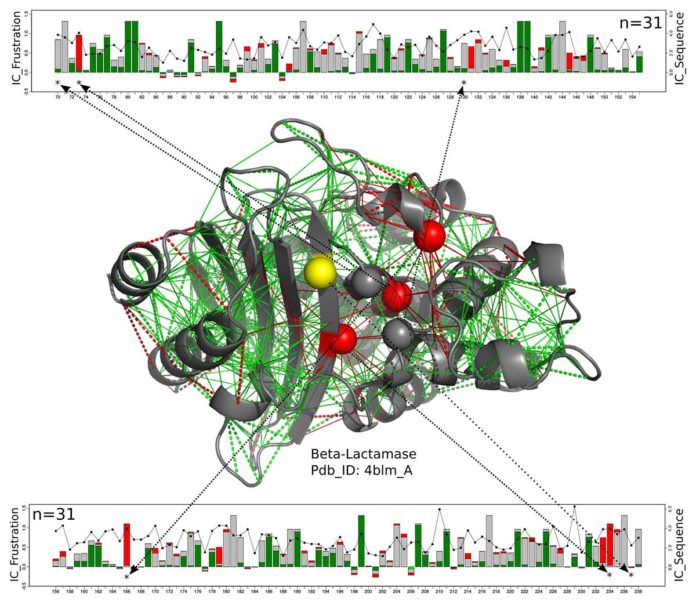
Scientists have studied different types of protein structures and discovered that whole families of enzymes share active site-frustrating signatures that extend to the second and third layers of amino acids, or shells, surrounding the enzyme.
Wolynes said, "The interesting thing we found is that the frustration usually exceeds the first shell. This means that there is a delicate but necessary subtlety to the functional constraints that require these three tanks to be properly designed. "
"We have seen signs of prolonged frustration in their efforts to modify the enzymes for new reactions. Sometimes they specifically changed the active site to catalyze different chemical reactions compared to natural reactions, but they found that amino acids far from the active site also needed to be modified for the enzyme to function effectively. "
"It was not obvious why, but models show that changes in the second and third shells improve the catalytic capacity of the enzymes. Our results have not been a total shock, but it is nice to see that prolonged frustration is quite common in nature and is a recurring theme in all known enzyme clbades. "
The study is detailed in the Proceedings of the National Academy of Sciences.
[ad_2]
Source link