[ad_1]
A recent study from France showed that the UK variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – also known as variant B.1.1.7 – can replicate much more efficiently in an epithelium reconstructed human bronchial tract, which may explain why it is spreading so rapidly in the human population. The study is currently available free of charge on the bioRxiv * pre-print server.
Since its emergence in 2019, circulating populations of SARS-CoV-2 have been known to acquire continuous genetic diversity. Therefore, within months, the D614G peak mutations were ubiquitous in all viral populations, however, with no evidence of a higher severity or mortality from coronavirus disease 2019 (COVID-19).
In September 2020, a variant named 20I / 501Y.V1 of the B.1.1.7 lineage emerged in the UK, with subsequent rapid spread to Western Europe and the US. There is ample evidence to show that this so-called ‘British variant’ can spread much faster and more efficiently compared to pre-existing European strains – especially in younger individuals.
In addition, new studies even show potentially increased mortality from the British strain, which, together with increased spread, can give rise to waves of emerging diseases. Nevertheless, salient biological evidence for different phenotypic characteristics compared to the original European variants is still lacking.
In this study, a research group from the University of Aix-Marseille in France (led by Dr Franck Touret) decided to assess the replication capacity of the British variant in different cell models, using a European strain ancestral D614G (line B1) for comparison. .
Highly specific / realistic cell models
In this study, the replication capacity of the viral variant 20I / 501Y.V1 (strain UVE / SARS-CoV-2/2021 / FR / 7b isolated in February 2021 in Marseille, France) was evaluated. in vitro and ex vivo, using strain B.1 BavPat D614G of the line that circulated in Europe in February / March 2020 for comparison.
The initial experiments were carried out in two cell lines frequently used for culturing SARS Cov-2: VeroE6 / TMPRSS2 (African green monkey kidney cells) and Caco-2 (i.e. human colorectal adenocarcinoma cells). ).
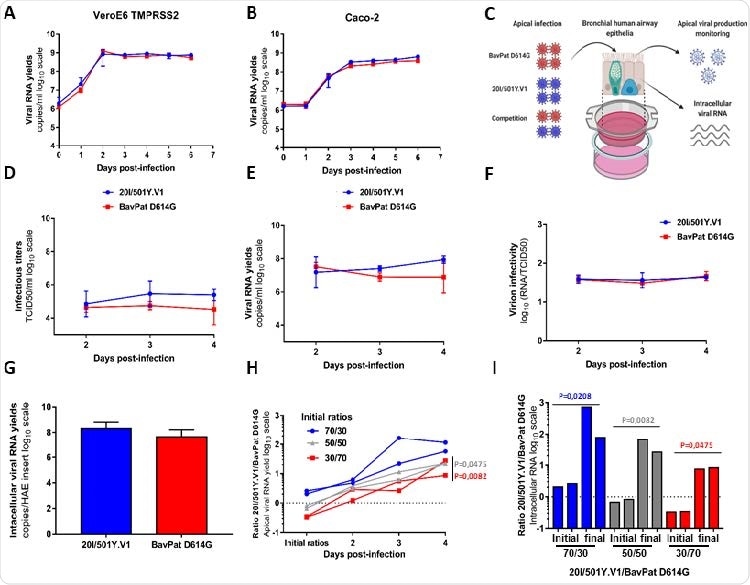
After revealing very similar replication kinetics, the researchers then assessed the replicative ability of the two strains using a model of reconstituted human airway epithelial cells of bronchial origin.
Specifically, after inoculating the epithelia from their apical side at a multiplicity of infection of 0.1 (which was done in order to mimic the natural infection route of infection), the researchers monitored the shedding of new virions from the apical side for 2-4 days. post infection and measured the intracellular yield of viral genetic material on day 4.
In vitro and ex vivo replication capacity of a 20I / 501Y.V1 variant in comparison with a B.1 D614G strain of the line. (AB) Replication kinetics in VeroE6 TMPRSS2 (A) and Caco-2 (B) cells. Viral replication was assessed using an RT-qPCR assay. (C) Graphical representation of experiments with reconstituted human airway epithelium (HAE) of bronchial origin. (DE) Kinetics of viral shedding on the apical side of the epithelium measured using a TCID50 149 assay (D) and an RT-qPCR assay (E). (F) Estimation of viral infectivities (i.e. the ratio of the number of infectious particles to the number of viral RNA). (G) Yield of intracellular viral RNA measured at 4 dpi using RT-qPCR assay. (AG) Data represent the mean ± SD of a triple. No statistical difference was observed between the two viral strains (p> 0.05; unpaired Mann-Whitney test). (H) Follow-up of the 20I / 501Y.V1 / BavPat D614G ratios on the apical side. Each row represents the results of an HAE insert. (I) Individual ratios 20I / 501Y.V1 / BavPat D614G estimated from intracellular viral RNAs at 4 dpi (I). The p (HI) values were determined relative to the initial ratios using the Kruskal-Wallis test followed by an uncorrected Dunn post-hoc analysis. The graphic representation was created with BioRENDER.
A new variety surpassing the old one
The study showed that the infectious titers and the yields of viral genetic material on the apical side of the cells were slightly higher for the British variant on days 3 and 4 of infection. The same was true for the intracellular yields of viral genetic material on day 4 of infection.
However, these differences were not significant, while the estimated relative infectivity of the virion (which was calculated as the ratio of the number of infectious particles to the number of viral RNA or genetic material) was similar for both. virus strains at all sampling times.
The highlight of this study was when the two viruses competed in a reconstituted human bronchial epithelium because the British variant subsequently defeated the ancestral strain – regardless of their initial ratio used in this study.
Understanding Strain Replacement
“Our results demonstrated that 20I / 501Y.V1 is in better shape than BavPat D614G in reconstituted human bronchial epithelium.”, say the authors of this study. bioRxiv paper. “This may be explained by the presence of the N501Y mutation in the receptor binding domain (RBD) of the spike protein, which enhances the binding of viral particles to the ACE2 receptor.”, they add.
Either way, it may bring some fitness benefits to the virus, as recently demonstrated in research projects using modified viral strains. More specifically, similar findings were seen with the D614G mutation, where the new G614 strains outperformed the original D614G strains when they were put into competition.
Everything says that this study could contribute to our better understanding of the gradual replacement of circulating strains by the SARS-CoV-2 20I / 501Y.V1 (or UK) variant. The emergence of different variants raises many questions about the future course of this pandemic. Therefore, more research efforts in this area are absolutely necessary.
*Important Notice
bioRxiv publishes preliminary scientific reports which are not peer reviewed and, therefore, should not be considered conclusive, guide clinical practice / health-related behaviors, or treated as established information.
Source link