.jpg)
[ad_1]
Researchers continue to better understand Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the virus that causes the ongoing coronavirus pandemic of 2019 (COVID-19). One such effort was reported in a new pre-print on the bioRxiv * server, dealing with the rollback phenomenon.
.jpg)
Going back in viral replication
Flashback refers to the retrograde movement of RNA-dependent RNA polymerase (RdRp), which is the crucial enzyme within the SARS-CoV-2 replication-transcription complex. This enzyme comprises three non-structural viral proteins sp7 / nsp82 / nsp12, and requires many other cofactors to perform its function. These include the nsp13 helicase and the nsp10 / nsp14 replay assembly.
Going back is important in the regulation of transcription via DdRp molecules. DdRp and the transcription machinery are associated with DNA recoil, but the RNA transcript makes its way back through the complex. This results in a single-stranded 3 ‘RNA transcript that is pushed through a secondary exit called the nucleoside-triphosphate (NTP) entry tunnel.
Objectives of the study
The current study attempts to solve a puzzle created by the putative structural arrangement of the nsp13 helicase bound to the virus’s replication-transcription complex (RTC). This involves both nsp13 and RdRp translocating on the RNA strand in opposite directions, the former in the 5 ‘-> 3’ direction and the latter in the 3 ‘-> 5’ direction on the same strand of ‘RNA template.
If the nsp 13 helicase is successful, it will push back the RdRp, in a reversible backward sliding motion. This backtracking has already been reported in DNA-dependent cellular RNA polymerases.
The researchers postulated that the helicase translocation mediates the rollback of RdRp in SARS-CoV-2 replication, making it energetically supportive. This process would allow RNA re-reading and template change during subgenomic RNA transcription.
Nsp 13 binds model RNA strand
They built RNA scaffolds based on the original SARS-CoV-2 RTC scaffold to test the RdRp assayed complexes (BTC). They found that the holo-RdRp binds to the RTC scaffold, but cannot effectively bind to the BTC scaffold without nsp13.
In the presence of nsp13, however, stable complexes formed. Using cryo-EM, they examined the nsp 13-RdRp-BTC assembly and found two major divergences from the nsp 13-RTC structures. First, a template-RNA-nsp 13 complex was observed, while second, a 3 ‘single-stranded p-RNA segment was extruded into the secondary NTP entry tunnel.
Engagement of the 5 ‘single stranded segment of tRNA with nsp 13 occurred in a seven nucleotide stretch between +14 and +8. A five-nucleotide stretch connected the t-RNA between the engaged nsp 13 and the RdRp in a disordered fashion.
Downgraded RNA extruded in an NTP entry tunnel
The discovery of the extruded segment in the NTP RdRp entry tunnel confirmed the presence of a BTC which is strongly analogous to the DdRp BTCs.
The researchers had previously solved the slit architecture of the DdRp active site, which was split into a channel for the DNA template strand and the NTP entry tunnel. Of these, one went above and the other went below the DdRp bridge propeller.
Similar to the bridge helix, the F motif of viral RdRp separates the two strands for BTC RNA. The presence of this entry tunnel allows an energetically favorable environment without steric barriers. This allows the retrograded RNA to leave the active site while avoiding obstructive polar interactions between RNA and protein.
This tunnel has an electrostatic surface containing positively charged arginine and lysine residues on the F motif, supplemented by the conserved residues of the RdRp C and E motifs.
NSP 13 promotes going back
SARS-CoV-2 wild-type holo-RdRp requires the nsp13 helicase for efficient binding to BTC scaffolds. This is not the case with the holo-RdRp containing nsp12 substituted with D760A.
Nsp12-D760 is a residue on the RdRp C unit which removes an essential magnesium ion from the catalytic complex. Magnesium is however absent from all RdRp structures without substrate such as BTC in this case.
Removing the D760 stabilizes the bond of the BTC to the scaffolding. This is because the original amino acid prevents tracking of the phosphate backbone of retrograded RNA without magnesium ions, according to the BTC structures suggested in the current study.
Alternatively, nsp 13 helicase is required to overcome this energy barrier. They found that the highest probability was that of a post-translocation state for RTC, with a 4-thio-U residue sequestered in the RNA-RNA hybrid. Although this makes it unavailable for crosslinking with nsp12, the addition of nsp 13 greatly increases crosslinking.
Bad incorporation favors going back
In addition, the study also shows that if a mismatched nucleotide is added to the growing strand of p-RNA at the 3 ‘end, it undergoes spontaneous unraveling to enter the RdRp NTP entry tunnel approximately 60% of the time, whereas a corresponding nucleotide spent 100% of the time in the pocket of the active catalytic site.
These results indicate the existence of a secondary tunnel to incorporate downgraded RNA, which is important for re-reading during RNA synthesis to ensure fidelity of transcription. This is analogous to cellular DdRps, although not evolutionarily related to them, indicating that this characteristic is essential for transcription enzymes.
Unpaired nucleotides spent more than 50% of their time unraveling from the template strand, either within or toward the NTP entry tunnel, likely preventing further translocation and stretching of the strand. This is further favored by the electrostatic and steric conditions of the NTP entry tunnel which encourage rollback.
Since translocation is prevented, the nsp 13 helicase can bind tightly to single-stranded tRNA, facilitating the helicase-mediated backtracking of the complex more easily via its 5 ‘-> 3’ translocation activity. .
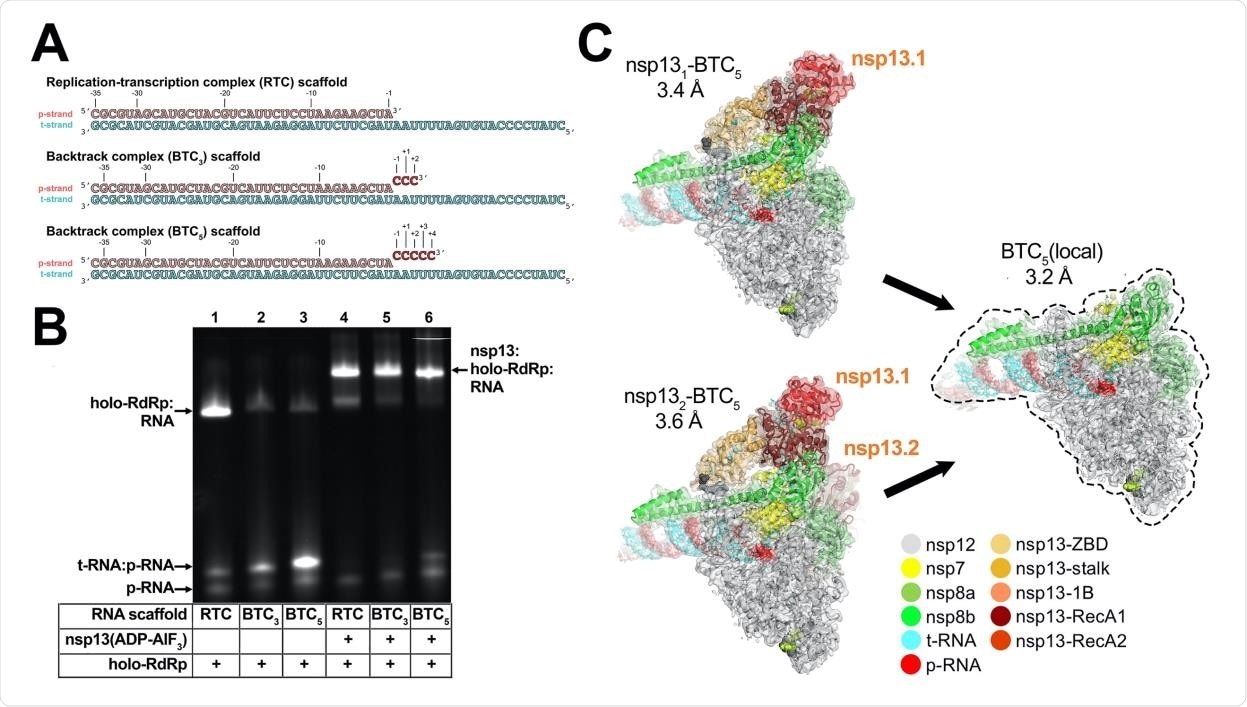
SARS-CoV-2 feedback complex. A. RNA scaffolds: (top) replication-transcription complex (RTC) scaffold (14); (bottom) complex backstroke scaffolding (BTC3 and BTC5). B. A native gel electrophoretic mobility shift assay reveals that holo-RdRp requires nsp13 (ADP-AlF3) to bind efficiently to BTC scaffolds. C. Cryo-EM structures of SARS-CoV-2 BTCs.
What are the implications?
The study thus validates an important prediction of the existing model of subgenomic transcription and re-reading, centered on the change of model. SARS-CoV-2’s ability to turn back the clock proves that pattern change can occur through this process.
Second, the incorporation of antivirals like remdesivir, i.e. nucleotide analogues, which are incorporated into the RNAs of the product by RdRps, could lead to a backtracking. Bad incorporation stops RdRp activity, so that nsp13 engages with the downstream template strand and initiates rollback.
Extrusion of the 3 ‘end of the pRNA from the NTP entry tunnel would then allow the viral replay components to break it down, and thus remove the mismatched nucleotide. This activity is essential for the resistance of coronaviruses to antivirals analogous to several nucleotides.
Understanding RdRp backtracking and its potential role in re-reading CoV can facilitate the development of therapies. “
*Important Notice
bioRxiv publishes preliminary scientific reports which are not peer reviewed and, therefore, should not be considered conclusive, guide clinical practice / health-related behaviors, or treated as established information.
Source link