[ad_1]
New study suggests that a new coronavirus test developed by scientists at the University of Washington and the Brotman Baty Institute for Precision Medicine produces faster and more sensitive results than conventional PCR tests for measuring severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA.
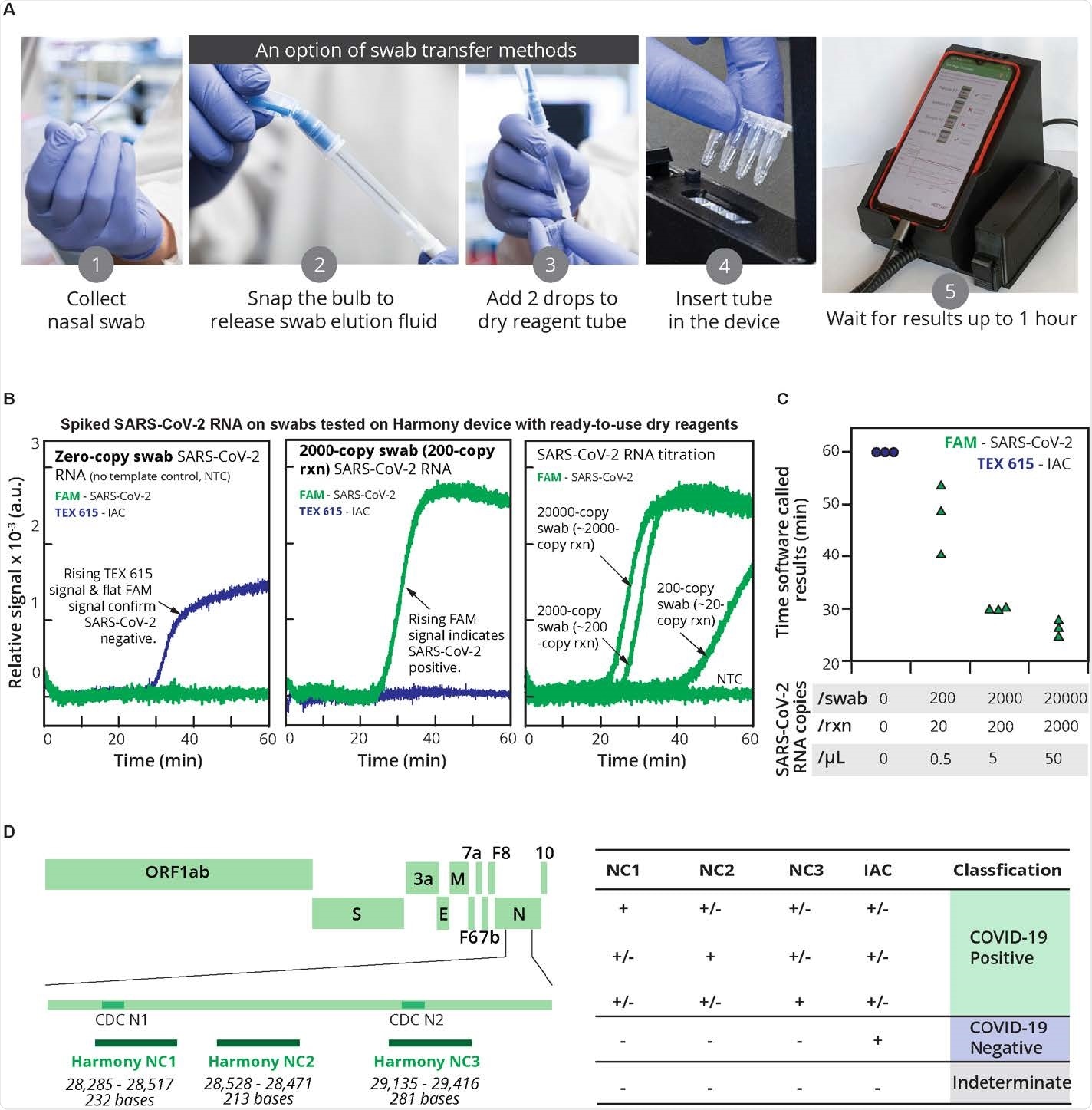
The RT-LAMP test kit known as “Harmony COVID-19” is a point-of-care test designed to be inexpensive and simple to use with the ability to examine up to four samples at a time. The device is intended to better help healthcare workers identify SARS-CoV-2 in patients.
Researchers are also hoping to expand Harmony COVID-19 to quickly test other respiratory infections, including seasonal SARS-CoV-2, influenza, and RSV.
The team writes:
“Tests such as Harmony will become standard practice to confirm the presence of viral infections that can be treated with antiviral therapies or that require infection control, as well as to help reduce the inappropriate use of antibiotics. In addition, the testing platforms developed for this pandemic could be more easily adapted to detect new diseases in order to increase preparedness for future pandemics. “
The study “Harmony COVID-19: a ready-to-use kit, a low-cost detector and a smartphone application for the detection of SARS-CoV-2 RNA at the point of care” is published on the preprint medRxiv* server.
How they did
The Harmony COVID-19 test kit was evaluated in the present study by asking healthcare workers to measure the RNA of artificial samples from the nasal and salivary matrix. In addition, healthcare workers also measured RNA from stored clinical samples.
New clinical samples could not be obtained in time for the study, which the authors admit to be a limitation of the study.
Healthcare workers learned how to use the test through written instructions and a video detailing the procedure.

Results suggest high precision and sensitivity in assay testing
Harmony’s COVID-19 testing has produced an inexpensive and simple rapid test with characteristics similar to conventional laboratory tests. Healthcare workers had 95% accuracy when using the test.
The results also showed the Harmony COVID-19 test kit to have high sensitivity.
The ready-to-use reagents in the test kit contributed to specificity and accuracy. The test detected approximately 15 copies / reaction of synthetic SARS-CoV-2 RNA and actual RNA samples collected at greater than 20 copies / reaction with no false positives. The test was also able to give results for high viral load samples within 17 minutes.
The test also successfully detected samples containing more than 20 viral particles in the human nasal matrix. In addition, the test had moderate sensitivity to saliva at similar concentrations.
Specifically, about 97% of the artificial samples in the nasal matrix were detected and 83% of the samples were detected in the saliva.
“Our current level of sensitivity of the test would be sufficient to detect SARS-CoV-2 in most nasal samples (~ 103 to ~ 109 copies / swab) or saliva (~ 104 to ~ 108 copies / mL) of infected individuals during the first week after onset, ”explained the research team.
The researchers attributed the high sensitivity and specificity of the assay to the use of a novel polymerase and probe design to specify between SARS-CoV-2 and IAC amplification.
Dosage improvements
The researchers point out that the test test could be modified to increase detection of SARS-CoV-2. They explain that performing the assay at a cooler and more optimal temperature for the enzyme reverse transcriptase followed by a higher temperature for DNA amplification could potentially increase the speed or sensitivity of detection of SARS- CoV-2.
Using an internal RNA amplification control that more accurately reflects the SARS-CoV-2 virus and that can be packaged in a viral envelope could also improve results. The current study used the Harmony COVID-19 test with an internal DNA amplification control, which fails to capture cDNA lysis and conversion processes.
“Advances in awareness and acceptance of large-scale infection testing, and the technology platforms developed will have a continued benefit in controlling COVID-19, reducing damage from other endemic diseases and fight against future pandemics, ”concluded the research team.
*Important Notice
medRxiv publishes preliminary scientific reports which are not peer reviewed and, therefore, should not be considered conclusive, guide clinical practice / health-related behavior, or treated as established information.
Source link