.jpg)
[ad_1]
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Wuhan, China, the virus has led to a global pandemic.
The emergence of several variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that have reduced the effectiveness of the vaccine and escaped neutralizing antibody therapies has caused much concern. Thus, it is imperative to develop strategies to inhibit all known and future variants of SARS-CoV-2.
According to new research by an international team of scientists, all variants of SARS-CoV-2, including the variants of concern (VOC) Alpha, Beta, Gamma, and Delta, exhibit an increased affinity for soluble recombinant human ACE2.
In addition, an important finding was that soluble ACE2 neutralized infection of VeroE6 cells and human lung epithelial cells by several strains of VOCs with markedly improved potency compared to reference isolates of SARS-CoV-2.
Two independent laboratories have confirmed the effectiveness of the inhibitor in inhibiting SARS-CoV-2 infections. The data show that the SARS-CoV-2 variants that have emerged around the world, including the current VOC and several variants of interest, are inhibited by soluble ACE2. This provides proof of principle for pan-SARS-CoV-2 therapy.
A preprinted version of this in vitro study, which is yet to be peer reviewed, is available on the bioRxiv* server.
.jpg)
Angiotensin converting enzyme 2 (ACE2)
When SARS-CoV-2 infects host cells, the viral protein Spike binds to the primary input receptor in human cells, the angiotensin-converting enzyme 2 (ACE2). This leads to the subsequent infection of the host cells.
Current COVID-19 vaccines induce the production of neutralizing antibodies that inhibit the Spike / ACE2 interaction. A therapeutic approach for the treatment of COVID-19 is the use of approved monoclonal antibodies that block the Spike / ACE2 interaction. Because of the importance of this interaction, the focus is on finding its molecular details.
Therefore, it is also one of the best validated drug targets for COVID-19 treatment. In this study, the scientists used a clinical grade recombinant human soluble ACE2 (APN01) which is already in phase 2 clinical trials.
Variants of SARS-CoV-2
Throughout the pandemic, several variants of SARS-CoV-2 have emerged. The World Health Organization has called some variants of VOCs because they are highly infectious and transmissible.
Many of these variants harbor mutations in the viral Spike protein that do not appear to affect the infectivity and transmissibility of the virus, but rather reduce the potency of vaccines and monoclonal antibody therapies.
They are also responsible for breakthrough infections. For example, the Delta variant can cause infections even in doubly vaccinated individuals.
This situation presents a need for universal strategies to prevent and treat current and future VOCs. To this end, scientists have explored the therapeutic potential of APN01.
APN01 test
Recombinant human clinical grade ACE2 was produced and used for this study. Spike protein receptor (RBD) binding domain and ACE2 binding studies were performed using ELISA and surface plasmon resonance analysis.
To test for neutralization of VOCs by APN01, neutralization assays were performed in VeroE6 cells and human lung epithelial cells. These neutralization tests were performed at the NIAID Integrated Research Facility at Fort Detrick, Frederick, MD, USA. To ensure data reproducibility, neutralization tests were repeated at the Karolinska Institutet, Stockholm, Sweden.
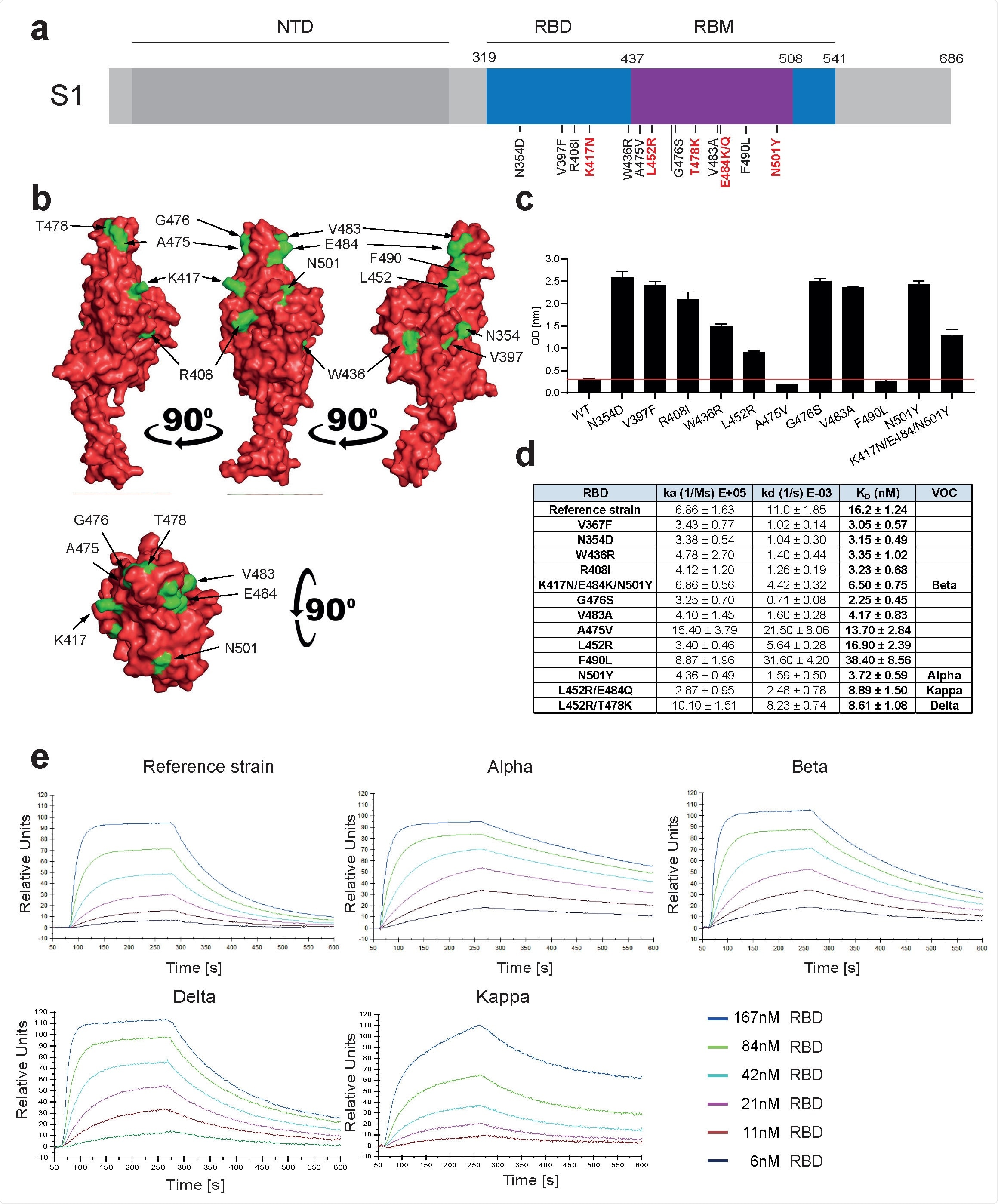
Increased affinity of APN01 interactions with SARS-CoV-2-RBD variants. (a) The diagram illustrates the structure of the S1 domain of the SARS-CoV-2 spike protein. The amino terminal domain (NTD), the receptor binding domain (RBD) in blue and in RBD the receptor binding motif (RBM) in purple are indicated. The numbers above represent the boundaries of the domain. Mutations within RBD / RBM are shown below along with the observed amino acid exchanges. In red, the mutations observed in the variants of concern (VOC). (b) PyMOL-rendered visualization of SARS-CoV-2 RBD. The render shows the SARS-CoV-2 RBD with the mutation sites indicated in green. (c) ELISA showing the binding strength of SARS-CoV-2 RBD carrying the mutations indicated at APN01. The axis labels indicate the substitutions of tested SARS-CoV-2 RBD variants. (d) Surface plasmon resonance analysis to derive the kinetic constants (ka, kd) and the affinity values (KD) of the SARS-CoV-2 RBD / APN01 interaction. The table lists both the variants tested and the amino acid substitution introduced as well as the designation of the respective variants of the mutations of concern tested in this study. The RBD strain reference sequence corresponds to the Wuhan SARS-CoV-2 isolate (e) Representative SPR sensor images for the SARS-CoV-2 RBD / APN01 interaction.
Blocks SARS-CoV-2 variants
The evolution of SARS-CoV-2 focuses on the Spike protein, in particular RBD. RBD mutations affect ACE2 binding. Clinical grade ACE2 showed a higher affinity for Spike RBD of the SARS-CoV-2 variants compared to Spike RBD of the original Wuhan SARS-CoV-2 isolate.
This increased binding affinity may be the reason for the increased infectivity of VOCs. Additionally, this increased affinity was also observed with full length Spike proteins.
Neutralization tests in VeroE6 cells with APN01 showed that APN01 neutralized all of the SARS-CoV-2 isolates tested. Most importantly, this inhibition was enhanced against all VOCs.
Equivalent results were obtained from a physiologically relevant cellular system, pulmonary epithelial cells. Interestingly, scientists observed that the neutralizing power correlated with the Spike / APN01 protein binding affinity.
In conclusion, ACE2 / APN01 showed strong binding to VOC RBDs or full length VOC Spike protein and strongly inhibited viral infection by SARS-CoV-2 isolates.
The neutralization tests have been independently validated at the Karolinska Institutet. These confirmatory experiments also showed that APN01 inhibited viral infection. Significantly, APN01 inhibited VOC infection with increased potency.
Limitations of this study
Scientists agree that this study has two limitations:
- This study used two different types of cells. Other cell types should also be studied to observe the same effect.
- If ACE2 is to be considered a universal agent, additional variants should be tested.
Universal agent
Current VOCs include Alpha, Beta, Gamma, and Delta variants, and interesting variants include Iota, Kappa, Eta, Mu, and Lambda variants.
New variations are expected due to large-scale vaccination programs. Some of these variants can lead to breakthrough infections and the rapid spread of the virus. Therefore, the design of universal therapeutic strategies is of paramount importance.
APN01 has been in a phase 2 trial in patients with severe COVID-19 using intravenous infusions. This test is now being extended to a larger patient population.
Scientists propose that the data from this study in conjunction with clinical data pave the way for the development of universal and pan-SARS-CoV-2 therapy.
*Important Notice
bioRxiv publishes preliminary scientific reports which are not peer reviewed and, therefore, should not be considered conclusive, guide clinical practice / health-related behavior, or treated as established information.
Source link