.jpg)
[ad_1]
In the year since the start of the 2019 coronavirus disease pandemic (COVID-19), there has been a huge investment in research into the virus that causes it, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS -CoV-2).
As such, numerous experimental studies have taken place to help unravel the structure and biochemistry of the globally dominant variant of this virus, strain D614G, as well as how this affects its function.
.jpg)
A new pre-print on the bioRxiv * The server aims to develop “a unified view and a working model consistent with the various experimental data”. The scientists used a computational approach to integrate the available data, and thus explore the molecular mechanisms of the D614 mutation. They applied atomistic modeling of the viral spike protein, conceiving it as an allosteric regulatory mechanism.
The peak protein
The SARS-CoV-2 tip has a C-terminal and N-terminal S1 and S2 subunit. The SI mediates virus-receptor attachment, with an N-terminal domain (NTD) and receptor binding domain (RBD). RBD is metastable and continually undergoes spontaneous conformational transformations, switching between the “down” and “up” positions. The peak can only bind to the receptor in the “up” conformation.
S2 mediates viral-host cell fusion. It is highly conserved and contains many peptides. The S1 is a dynamic and protective shield of the fusion mechanism.
Upon viral binding to the host angiotensin converting enzyme receptor 2 (ACE2), the spike protein is cleaved at the S1 / S2 interface, and the two subunits dissociate. This triggers a series of conformational changes that take place in the fusion of viral and cell membranes.
Study objectives
The study aimed to simulate the structure of native and mutant SARS-CoV-2 spike proteins. To this end, the researchers used large-scale simulations, as well as analyzes of protein stability and dynamic fluctuation communication, and community network analysis.
The D614G mutation subtly disrupts collective movements
Previously, some scientists explained the increased infectivity of the D614G mutant as being due to the altered conformational dynamics of the peak, with open conformation being favored – the “open” hypothesis. This would allow the RBD peak to make contact with the host ACE2 receptor.
This level of alteration was not observed in the present study. The use of CG-CABS simulations followed by atomistic reconstruction shows that the D614G mutation has conformational dynamics very similar to wild type in the closed and open states. The effect of the mutation is therefore very delicate, comprising a large number of small localized changes distributed throughout the structure of the protein. These affect both inter-protomeric and intra-protomeric interactions, preferentially regulating certain collective movements in closed conformation.
A closer look at how collective movements and rearrangements occur after the D614G mutation may shed more light on how this affects infectivity.
D614G modulates stability and propensity for allosteric communication
The researchers also looked at the stability and communication propensities of amino acids both in the wild type and in the mutant spike protein of the virus. They found that the D614 site, as well as the Q613 site, anchor and incorporate major residues in regulatory or hinge centers that regulate global peak movements as well as allosteric changes in tip-to-state structure. closed and open.
These articulation centers are energetic hotspots, with intra- and inter-protomeric contacts occurring near these locations, primarily in the rigid S2 subunit. D614G causes the greatest change in energy compared to other similar substitutions, with stabilization of closed and open tip conformations.
The mutation stabilizes the peak trimer while reducing premature shedding of the S1 domain – the “S1-shedding” hypothesis. This increases the number of functional peaks and the infectivity of the variant.
The D614G mutant, however, disrupts the hinge clusters and alters functional movements in the trimer, which may increase the willingness of tip protomers to switch to the open conformation. Thus, the stabilizing effect of this mutation differs in the closed and open states. This may help to unify the “opening” and “S1 shedding” hypotheses explaining the effect of this mutation.
Improved allosteric signaling in the open state
Community network analysis of SARS-CoV-2 peak proteins demonstrates the ability of this mutation to increase the number of stable communities in open conformation, and allosteric centers for S1-S2 interdomain interactions.
Their results show that the mutation is capable of stimulating the long-range signaling of the peak, by reorganizing allosteric interactions in the closed state. This increases stability and communication at the S1 / S2 interface, with better allosteric signaling between the two domains in the open state.
At the same time, the mutation restricts S1 mobility, reducing its loss, while improving the thermodynamic advantages of the open state. As a result, RBD and NTD are potentially more exposed to the host receptor, which promotes increased infectivity.
This supports the role of the mutation in promoting the open state by optimizing allosteric signaling in this state and limiting the excretion of S1.
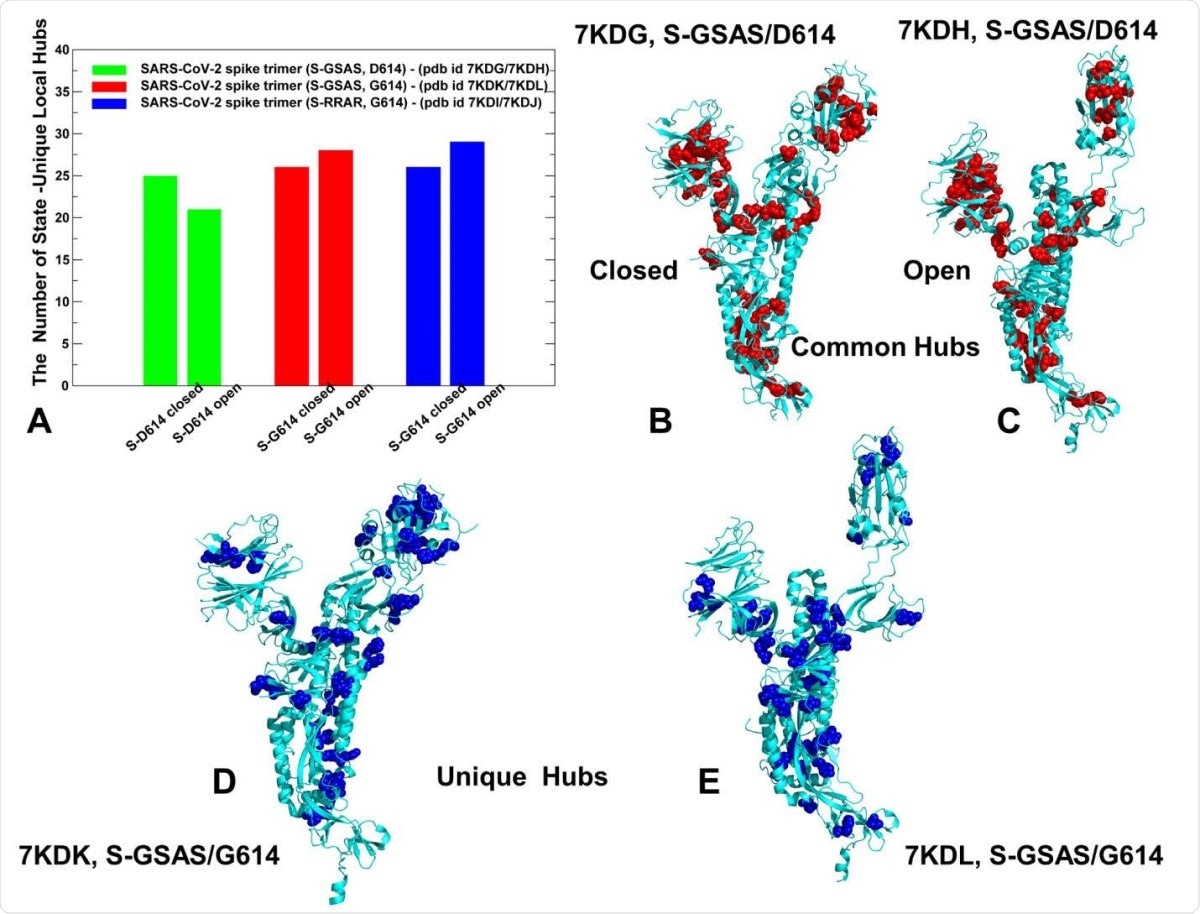
Network hub analysis of SARS-CoV-2 S-D614 and S-D614G mutant structures. (A) The quantitative assessment of the number of unique hubs in closed and open forms of SGSAS / D614 (indicated in green bars), closed and open forms of S-GSAS / G614 (red bars) and SRRAR / G614 ( blue bars)). The structural mapping of common communities shared by closed and open states is projected onto a single protomer for S-GSAS / D614 in closed state all closed (B) and S-GSAS / D614 in open state (C) . The hubs are represented by red spheres. (D) Structural mapping of the unique in the closed form of S-GSAS / G614 (pdb id 7KDK). (E) Structural mapping of single concentrators for the S-GSAS / G614 in the open state (pdb id 7KDL). The hubs are shown in blue spheres. The mapping is projected onto a single protomer represented in cyan ribbons.
What are the implications?
The study suggests the allosteric regulatory function of the D614G mutation, acting on both adjacent and distant sites. The results show that the D614 residue is that which controls transitions of the receptor binding domain (RBD) of the viral spike protein between the closed state and the open state. The D614G mutation can improve peak stability in both open and closed forms, but it allows the open form to be more thermodynamically favorable.
As a result, say the researchers, this mutation is associated with reduced shedding of the S1 domain of the spike protein. This in turn results in the higher infectivity of this strain.
The results reveal that the D614G mutation reduces S1 excretion, affecting local interactions in the spike protein. The main effect continues to be via its allosteric effects on peak stability and communications within protein residue networks.
This model of allosteric regulation could help explain how the D614G mutation causes its effects. In the future, this could help explain how mutations allow immune evasion. Some recent studies show that the presence of D614G has a striking stimulating effect on mutations in remote areas. These mutations in turn lead to immune leakage.
Taking into account the functions of pike proteins from SARS-CoV-2 through the prism of an allosterically regulated machine may prove useful in discovering functional mechanisms and rationalizing the growing body of diverse experimental data via allosteric models. underlying the signaling events.
Its wide impact could lead to a global amplification of the effects of other mutations, explaining its rise to world domination. Further studies on the advanced protein-mediated centers will help to understand the mechanisms of viral infectivity as well as to identify and develop appropriate inhibitors, to effectively treat this infection.
*Important Notice
bioRxiv publishes preliminary scientific reports which are not peer reviewed and, therefore, should not be considered conclusive, guide clinical practice / health-related behaviors, or treated as established information.
Source link