
[ad_1]
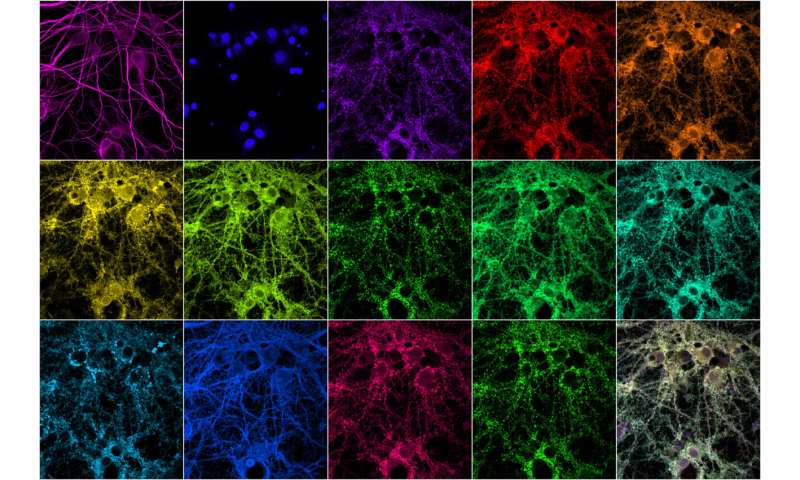
MIT engineers have developed a technique that allows them to quickly image many different proteins within a synapse. Bottom right is a composite of the other images. Credit: Syuan-Ming Guo and Li Li
Our brain contains millions of synapses, connections that transmit messages from neuron to neuron. Hundreds of different proteins are present in these synapses, and their dysfunction can lead to problems such as schizophrenia and autism.
Researchers at MIT and the Broad Institute of Harvard and MIT have now developed a new way to rapidly image these high-resolution synaptic proteins. With the help of fluorescent nucleic acid probes, they can mark and image an unlimited number of different proteins. They demonstrated the technique in a new study in which they imaged 12 proteins in cell samples containing thousands of synapses.
"Multiplexed imaging is important because there is a great deal of variability between synapses and cells, even within the same brain," says Mark Bathe, an associate professor of biological engineering at MIT. "You really need to look at the sample proteins simultaneously to understand what the subpopulations of different synapses look like, to discover new types of synapses, and to understand how genetic variations affect them."
The researchers plan to use this technique to study the fate of synapses when they block the expression of genes associated with specific diseases, hoping to develop new treatments that can reverse these effects.
Bathe and Jeff Cottrell, director of translational research at the Stanley Center for Psychiatric Research at the Broad Institute, are the lead authors of the study, which appears today in Nature Communications. The main authors of the document are former post-docs Syuan-Ming Guo and Remi Veneziano, a former graduate student Simon Gordonov and a former research scientist Li Li.
Imaging with DNA
Synaptic proteins have various functions. Many of them contribute to the formation of synaptic scaffolds, involved in the secretion of neurotransmitters and the processing of incoming signals. While synapses contain hundreds of these proteins, conventional fluorescence microscopy is limited to imaging up to four proteins at a time.
To increase this number, the MIT team has developed a new technique based on an existing method called DNA PAINT. Using this method, originally designed by Ralf Jungmann of the Max Planck Institute of Biochemistry, researchers tag proteins or other molecules of interest with a DNA-antibody probe. Then, they image each protein by delivering an "oligo" fluorescent DNA that binds to DNA-antibody probes.
DNA strands intrinsically have low affinity between them. They bind and disassemble periodically, creating a flashing fluorescence using super-resolution microscopy. However, the imaging of each protein takes about half an hour, which makes it impractical for imaging many proteins in a large sample.
Bathe and his colleagues decided to create a faster method to analyze a large number of samples in a short time. To achieve this, they modified the DNA-dye imaging probe so that it binds more closely to the DNA-antibody, using what's called locked nucleic acids. . This gives a much brighter signal, so that imaging can be performed faster, but with a slightly lower resolution.
"When we do 12 or 15 colors on a single neuron well, the whole experiment takes an hour, compared to a night for the equivalent of super-resolution," says Bathe.
The researchers used this technique to tag 12 different proteins present in the synapse, including scaffold proteins, cytoskeleton-associated proteins, and proteins known to label excitatory or inhibitory synapses. One of the proteins studied is the Shank3, a scaffold protein that has been linked to autism and schizophrenia.
By analyzing protein levels in thousands of neurons, researchers have been able to identify groups of proteins that tend to associate with each other more often than others and learn how different synapses vary. in the proteins they contain. This type of information could be used to help classify synapses into subtypes that can help reveal their functions.
"Canonical synaptic types are inhibitory and excitatory, but it is assumed that there are many different subtypes of synapses, with no real consensus on what they are," Bathe says.
Understanding the disease
The researchers also showed that they could measure changes in synaptic protein levels after neuronal treatment with a compound called tetrodotoxin (TTX), which strengthens synaptic connections.
"By using conventional immunofluorescence, you can usually extract information from three or four targets of a single sample, but with our technique, we have been able to extend that number to 12 different targets." in the same sample, we applied this method to examine synaptic remodeling, occurs after TTX treatment, and our results corroborate previous work that revealed coordinated synaptic protein regulation after TTX treatment, "says Eric Danielson, postdoc at MIT and author of the study.
Researchers are now using this technique, called PRISM, to study how the structure and composition of synapses are affected by the inactivation of genes associated with various disorders. Sequencing the genomes of people with disorders such as autism and schizophrenia has revealed hundreds of genetic variants linked to the disease and, for most of them, scientists have no idea of how they contribute to the disease.
"Understanding how genetic variation influences the development of neurons in the brain, as well as their synaptic structure and function, is a major challenge for neuroscience and understanding the genesis of these diseases," said Bathe.
A step closer to finding a cure for brain diseases
Quote:
One technique allows imaging of individual proteins in synapses (September 26, 2019)
recovered on September 26, 2019
on https://medicalxpress.com/news/2019-09-technique-image-individual-proteins-synapses.html
This document is subject to copyright. Apart from any fair use for study or private research purposes, no
part may be reproduced without written permission. Content is provided for information only.
[ad_2]
Source link