
[ad_1]

Scientists used an adaptation of CRISPR genome editing technology to develop a new peptide display platform that can be used as a tool to identify antibodies in patient blood samples. Researchers at Howard Hughes Medical Institutes and Harvard Medical School suggest that the new technology, which they called PICASSO (Cas9-Mediated Self-Organizing Peptide Immobilization), could inspire a new class of medical diagnostics and launch a host of other applications.
For their study reported in Molecular cell, which is titled “CRISPR-based Peptide Library Display and Programmable Microarray Self-assembly for Rapid Quantitative Protein Binding Assays,” the researchers used the platform to detect the derived protein-binding antibodies of pathogens, including SARS-CoV-2 from the blood to recover patients with COVID-19. The work was led by Stephen Elledge, PhD, at Harvard Medical School and Brigham and Women’s Hospital. The first author of the article, Karl Barber, PhD, is a 2018 Schmidt Science Fellow, with much of the technology development work taking place during his research internship in the laboratory of corresponding author Elledge.
CRISPR-Cas9 technology has been adapted for a range of applications in precise genome editing and transcriptional regulation, allowing scientists to efficiently manipulate genetic sequences “at will,” the authors noted. But the ability of different Cas nuclease enzymes to recognize and respond to nucleic acid sequences complementary to their linked single-guide RNAs (sgRNAs) has also “inspired further the development of in vitro technologies based on CRISPR,” including a point fast service. identification of pathogens and smart hydrogels sensitive to nucleic acids. “CRISPR-inspired systems have been widely developed for applications in genome editing and nucleic acid detection,” they noted.
The PICASSO technology developed by Barber and colleagues uses a modified Cas9 enzyme to facilitate the study of custom peptide libraries and overcome the limitations of current display technologies. “By developing PICASSO, we have demonstrated the self-assembly of a library of multiplexed peptides using a CRISPR-based system, making rapid personalized protein studies possible in any laboratory with access to common molecular biology reagents, “they wrote. The PICASSO approach exploits customizable collections of proteins, which are attached to a catalytically inactive Cas9 enzyme (dCase9). This variant will bind to DNA, but not cut it, as it would for genetic modification applications. Peptide collections are fused to dCAS9 and bar coded with unique unique guide RNA (sgRNA) sequences.
These dCas9-peptide fusions then self-assemble at positions on a DNA chip surface complementary to their sgRNA barcodes. So when applied to a microchip containing thousands of unique DNA molecules, each protein in the mixture will self-assemble at the position on the chip containing its corresponding DNA sequence. “… the unique mixed pool of dCas9 fusion peptides is capable of locating user-programmed positions on a microarray surface containing DNA sequences complementary to the sgRNA barcode of each peptide,” noted the team. The resulting DNA template self-assembling peptide arrays can then be used for large-scale protein studies. “The dCas9 fusion display and self-assembly microarray construction via PICASSO bypasses many caveats of other display platforms, making custom peptide library studies faster and more widely accessible,” investigators said.
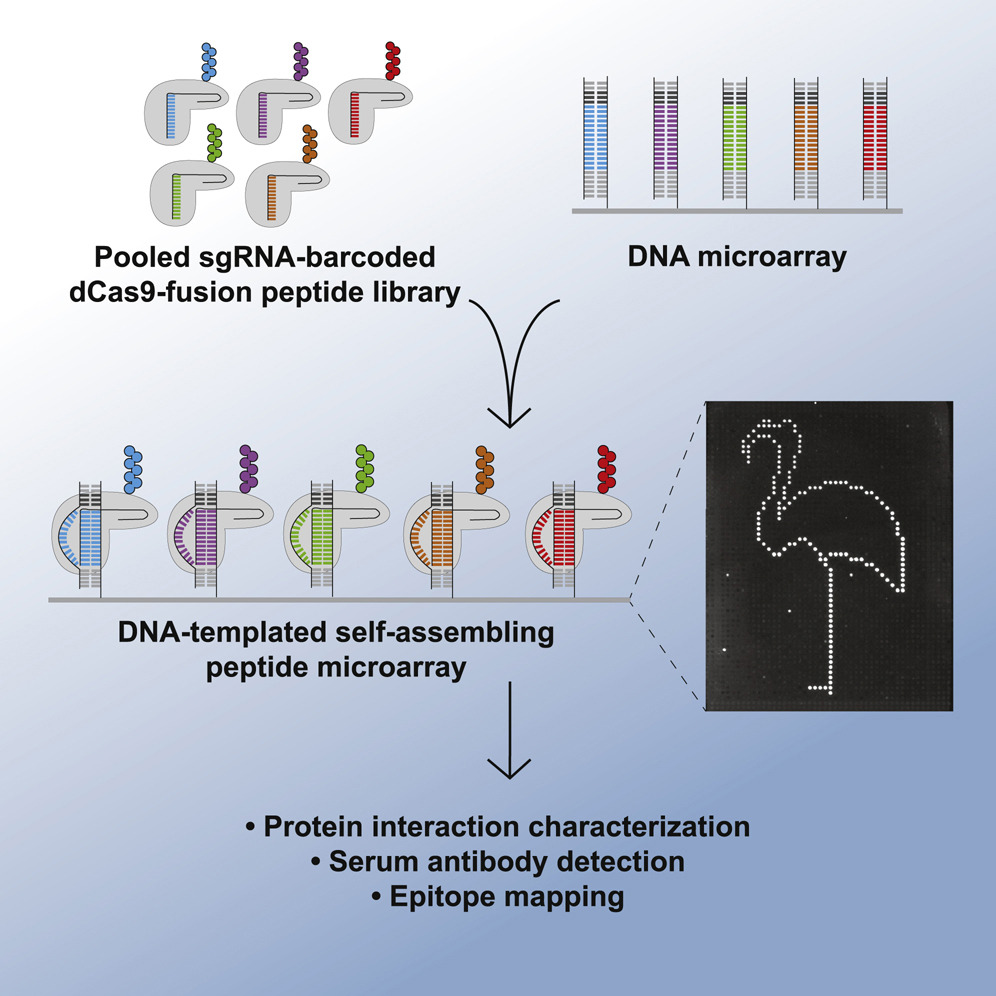
Describing the technology, Barber said, “Imagine you wanted to paint a picture on canvas, but instead of painting in the normal way, you mix all of your paints together, splash them onto the canvas and the perfect image emerges. With our new technique, you place DNA molecules in defined locations on a surface and each protein in a mixture will then self-assemble to its corresponding DNA sequence, like an automated paint-by-number kit. The resulting DNA matrix protein microarrays allow you to quickly identify antibodies in clinical samples that recognize all the proteins of interest to you. Thus, by applying a blood sample to the PICASSO chip, the chip proteins that are recognized by the patient’s antibodies can be identified.
Barber commented: “In this work, we have demonstrated the application of PICASSO for protein studies, creating a tool that we believe could be quickly adapted for medical diagnosis. Our protein self-assembly technique could also be exploited for the development of new biomaterials and biosensors simply by attaching DNA targets to a scaffold and allowing Cas9-bound proteins to bind. The authors further asserted: “Without next-generation sequencing requirements or multiple rounds of selection, PICASSO can be performed within a few hours after applying the dCas9-sgRNA library to a ds DNA chip (compared to several days or weeks) and avoids sequencing the costs and PCR artifacts associated with nucleic acid-based displays.
Group leader Elledge commented: “One of the most exciting aspects of this work is demonstrating how CRISPR can be applied in an entirely new setting. Previously, CRISPR was primarily used for gene editing and detection of DNA or RNA. PICASSO brings the power of CRISPR into a new field of protein studies, and the molecular self-assembly strategy we show may help develop new research and diagnostic tools.
And going forward, the team noted: “… The CRISPR and PICASSO-based protein display can also be developed for classes of proteins that are generally incompatible with other displays… We anticipate that the display based on on dCas9 and PICASSO will be useful for studying custom peptide and protein libraries for many additional applications, including multiplexed diagnostics, discovery of enzyme substrates, and protein evolution and design experiments.
[ad_2]
Source link