[ad_1]
In recent years, researchers have used DNA to encode everything from the operating system to malware. Rather than being a technological curiosity, these efforts were serious attempts to take advantage of the properties of DNA for long-term data storage. DNA can remain chemically stable for hundreds of thousands of years, and we’re unlikely to lose the technology to read it, which you can’t say about things like ZIP drives and MO disks.
But until now, writing data to DNA has involved converting the data into a basic sequence on a computer, and then ordering that sequence from a location that operates a chemical synthesizer – living things. do not actually fit into the picture. But separately, a group of researchers had sought to record biological events by modifying a cell’s DNA, allowing them to read the history of the cell. A group at Columbia University has now figured out how to merge the two efforts and write data to DNA using the voltage differences applied to living bacteria.
CRISPR and data storage
The CRISPR system was developed to edit genes or cut them completely from DNA. But the system first caught the attention of biologists because it inserted new sequences into DNA. For full details, check out our Nobel coverage, but for now, know that part of the CRISPR system is to identify the DNA of viruses and insert copies of it into the bacterial genome in order to recognize it if the virus reappears. .
The Columbia group figured out how to use it to record memories in bacteria. Let’s say you have a process that activates genes in response to a specific chemical, like a sugar. The researchers hijacked this to also activate a system that makes copies of a circular piece of DNA called a plasmid. Once the copy number was high, they activated the CRISPR system. Given the circumstances, it was most likely to insert a copy of the plasmid DNA into the genome. When sugar was not present, it usually inserted something else.
Using this system, it was possible to tell if a bacterium was exposed to sugar in its past. It’s not perfect, because the CRISPR system doesn’t always insert something when you want it, but it works on average. So you just need to sequence enough bacteria to determine the average sequence of events.
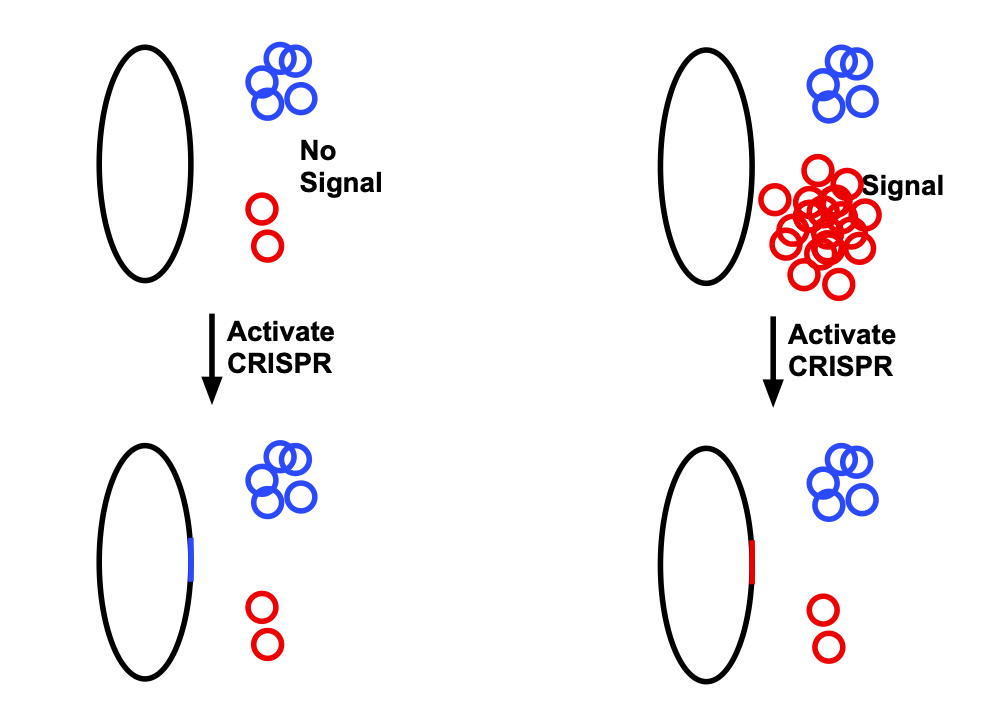
To adapt this to data storage, the researchers used two plasmids. One is the same as described above: present at low levels when a specific signal is absent, and present at very high levels when the signal is around. The second is still present at moderate levels. When CRISPR is activated, it tends to insert sequences of any plasmid present at higher levels, as shown in the diagram below.
John timmer
On its own, this only stores one bit. But the process can be repeated, creating a stretch of DNA which is a series of inserts derived from the red and blue plasmids, the identity being determined by the presence or absence of the signal.
Give it a jerk
It’s a neat system but far enough removed from the kinds of things we normally associate with producing data – the output of a sensor reading or calculation is rarely a sugar or an antibiotic mixed with a bunch of bacteria. Getting bacteria to react to an electrical signal has proven to be relatively straightforward. E. coli is capable of altering the activity of genes depending on whether it is an oxidizing or reducing chemical environment. And the researchers could alter the environment by applying voltage differences to a specific chemical in the culture with the bacteria.
Specifically, the voltage difference would change the oxidative state of a chemical called ferrocyanide. This in turn caused the bacteria to alter the activity of the genes. By designing the plasmid to respond to the same signal as these genes, the researchers were able to control plasmid levels by applying different voltages. And they could then record this level of this plasmid by activating the CRISPR system in these cells.
It’s pretty easy to see how each of the inserts in a series can be considered a zero or a one, depending on the identity of the insert. But remember that this system is not perfect; quite regularly, CRISPR would not insert anything when enabled, which would shift all subsequent bits. Because this process is random, the longer the series of bits you are trying to encode, the more likely it is that at least one of them will end up being ignored.
To limit this problem, the researchers kept their data at three bits per bacterial population. Even so, they had to train a supervised learning algorithm to reconstruct the most probable bit series based on an average of the sequences found in the population. And, even with that, the system failed to recognize the string of bits about six percent of the time. In the end, they decided to use a parity bit that was the sum of the first two to allow for error correction, and then edited many populations in parallel.
(By giving the plasmids in each population a unique sequence tag called a ‘barcode’, it was possible to mix many of them into a single population after the bits were encoded and unravel everything after the DNA sequence.)
With everything in place, they managed to store and read “Hello world!” They even put the bacteria in potting soil for a week and showed they were able to pick up the message. (Keeping them in the freezer obviously works best.) They estimate that the message can be stored for at least 80 generations of bacteria.
Let’s be clear: as a storage medium, in its current form, it’s pretty terrible. If you wanted to put data into DNA, you had better have the DNA chemically synthesized. But it’s fascinating to think that we could pass electrical signals directly to altered DNA, and there may be ways to improve the system now that it’s been established.
Nature Chemical Biology, 2021. DOI: 10.1038 / s41589-020-00711-4 (About DOIs).
[ad_2]
Source link