[ad_1]
Scientists at the Chalmers University of Technology in Sweden claim to have refuted the current theory of how DNA binds. As is generally believed, it is not hydrogen bonds that bind both sides of the DNA structure. Instead, water is the key. The paper describing their discovery ("Hydrophobic Catalysis and the Potential Biological Role of DNA De-piling Induced by Environmental Effects"), which could pave the way for a new understanding of medical research and life sciences , appears in PNAS.
"The stacking of hydrophobic bases is a major contributor to the stability of the double helix of DNA. We report the discovery of specific effects of stacking in some semi-hydrophobic environments. Water miscible ethylene glycol ethers have been found to modify the structure, dynamics and reactivity of DNA by mechanisms possibly related to biologically relevant hydrophobic catalysis. Spectroscopic data and optical clamp experiments show that the base stacking energies are reduced while the hydrogen bonds of the base pairs are enhanced. We propose that a modulated water chemical potential can promote "longitudinal breathing" and the formation of unstacked holes while dissociation of bases is suppressed, "the investigators wrote.
"Linear flow dichroism in 20% diglyme indicates a 20-30% decrease in DNA persistence length, supported by increased flexibility in single-molecule nanochannel experiments in polyethylene glycol." . Limited hyperchromicity (3-6%), but unaffected circular dichroism is consistent with transient unstacking events while maintaining an overall mean conformation of B-DNA. Additional information on depilatory dynamics is obtained from the binding kinetics of large ruthenium complexes interspersing the wires, indicating that the hydrophobic effect provides a DNA unstacking frequency of 10 to 100 times higher and a population of "open holes" of the order of 10-2 against 10-4 in normal aqueous solution.
"The polyethylene glycol-catalyzed spontaneous exchange of DNA strands leads us to propose that the hydrophobic residues in the L2 loop of the RecA and Rad51 recombination enzymes can help gene recombination via the modulation of chromosomes." 39, water activity near the helix of the DNA by hydrophobic interactions, as described here. We assume that such hydrophobic interactions can also play a catalytic role in other biological contexts, such as in polymerases.
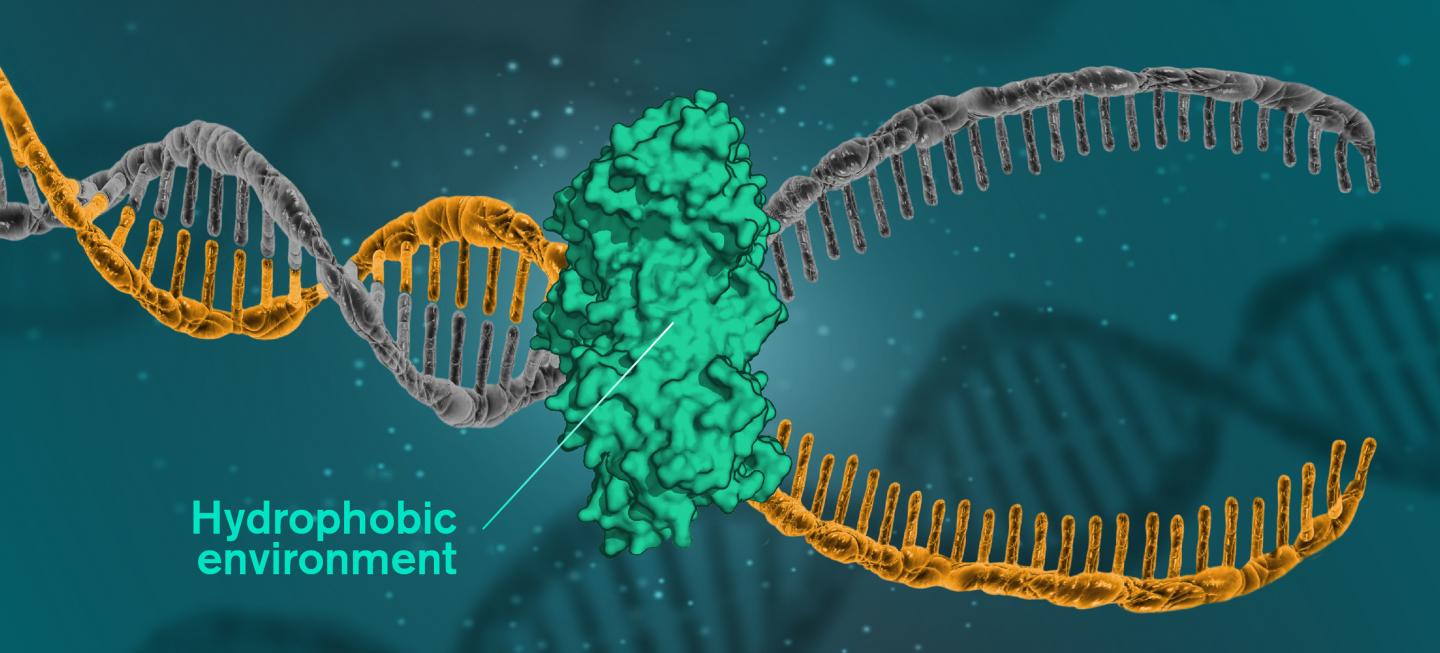
The research team stated that the secret of the helical structure of DNA may lie in the fact that the molecules have a hydrophobic interior, in an environment consisting mainly of water. The environment is therefore hydrophilic, while the nitrogenous bases of the DNA molecules are hydrophobic, repelling the surrounding waters. When the hydrophobic units are in a hydrophilic environment, they group together to minimize their exposure to water.
The role of hydrogen bonds, previously considered crucial for the consolidation of DNA helices, seems to be more related to the sorting of base pairs, so that they bind in the right order.

The discovery is crucial to understanding the relationship between DNA and its environment, said Bobo Feng, PhD, one of the researchers behind the study and postdoctoral fellow at Chalmers University of Technology.
"The cells want to protect their DNA and not expose it to hydrophobic environments, which can sometimes contain harmful molecules," Feng said. "But at the same time, the cells' DNA must open up to be able to be used. We think that the cell retains its DNA in an aqueous solution most of the time, but as soon as a cell wants to use something with its DNA, like reading it, copying it or repairing it, it exposes the DNA to a hydrophobic environment. "
Reproduction, for example, implies that the base pairs dissolve and open. The enzymes then copy both sides of the helix to create a new DNA. When it is a question of repairing the damaged DNA, the damaged areas are subjected to a hydrophobic environment, to be replaced. A catalytic protein creates the hydrophobic environment. This type of protein is at the heart of all DNA repair, which means it could be the key to fighting many serious diseases.
Understanding these proteins could provide a lot of new information on how we could, for example, fight resistant bacteria, or even potentially cure cancer, according to scientists. Bacteria use a protein called RecA to repair their DNA, and researchers believe their findings could provide new insights into how this process works, potentially offering methods to stop them and kill the bacteria.
In human cells, the Rad51 protein repairs the DNA and fixes the mutated DNA sequences, which might otherwise cause cancer.
"To understand cancer, we need to understand how DNA is repaired. To understand this, we must first understand the DNA itself, "Feng continued. "Until now, we have not done it because we thought hydrogen bonds were what held him. We have now shown that it is rather hydrophobic forces. We have also shown that DNA behaves totally differently in a hydrophobic environment. This could help us understand the DNA and its repair. Nobody had previously placed DNA in a hydrophobic environment like this one and studied its behavior. No wonder no one has discovered it so far. "
[ad_2]
Source link