[ad_1]
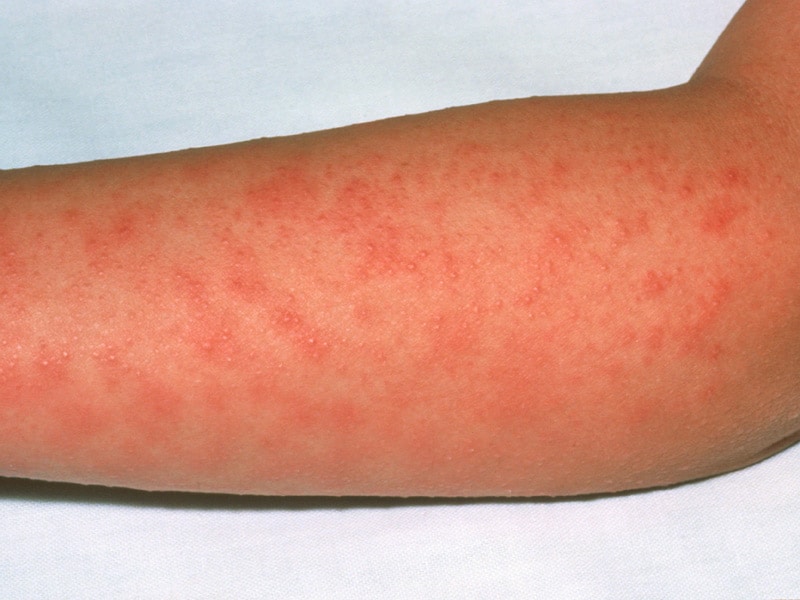
A new strain of the group A streptococcus bacteria called M1UK, related to scarlet fever and sepsis has been identified in England and Wales.
The discovery was made by researchers led by Imperial College London who reported their findings in the newspaper. Infectious diseases lancet newspaper.
Professor Shiranee Sriskandan, lead author of the Infectious Diseases Department at Imperial Oil, Medscape News UK"In a nutshell, an already dominant bacterial strain has already changed in a few small genetic regions … and the only biological change we can observe in the virus is that it produces more of a particular type of toxin."
So far, cases seem to be limited in the UK, but study authors have written that their discovery "underscores the need for global surveillance and increased vigilance ".
Scarlet fever epidemic
Significant increases in the number of cases of scarlet fever have been observed in recent years: more than 15 000 cases in 2014, more than 17 000 in 2015 and more than 19 000 in 2016.
Invasive infections caused by the same bacterium, Streptococcus pyogenes (S pyogenes), also increased in 2016 compared to the previous 5 years.
Cases were evaluated by emm genotypes.
The initial increase in the number of cases of scarlet fever in 2014 was associated with the types of emm3 and emm4 strains of streptococcus A. However, in 2015-2016, emm1 strains became dominant in throat infections.
The emm1 strains have also become more dominant in England and Wales.
In the spring of 2016, 42% of the invasive strains collected were emm1, compared to 31% the previous year.
The researchers explain that the new strain of Streptococcus pyogenes could explain the increase in the number of invasive cases.
Professor Sriskandan told us: "The strain that emerged is related to this previously dominant strain called emm1, and it changed very little in that it acquired 27 fairly small mutations. Having done so, she has been very successful. He seems to have had the ability to develop within the population by causing both throat infections and the much rarer invasive infection.
"What is interesting, from a scientific point of view, is that everything that has happened to this bacteria has made it more" fit "or has a better ability to cause throat infections in a population.
"One of the ways in which it has manifested itself biologically is that it can produce more of a type of toxin called SpeA, or scarlet fever toxin."
Sequencing of the genome
The new strain type was identified after sequencing the genomes of all 135 non-invasive emm1 isolates collected in northwestern London between 2009 and 2016. The same was true for 552 invasive isolates collected in England and Wales during the seasonal rise of the disease from 2013. 2016
These were compared and evaluated to determine the production of toxins by different emm1 strains.
Most of the emm1 strains in 2015 and 2016 were found to be phylogenetically distinct, which the researchers called M1UK.
This M1UK clone produced nine times more streptococcal A pyrogenic exotoxin (SpeA) than other emm1 strains (190 ng / mL versus 21 ng / mL).
It was found that this strain was circulating in England as early as 2010.
In 2016, M1UK accounted for 84% of all emm1 genomes analyzed from cases in England and Wales.
According to Professor Sriskandan, the M1UK streptococcal type can also be treated with penicillin and alternatives.
An analysis was also performed on 2800 cases of Strep A genome sequences from around the world. Unique isolates of M1UK have been found in the United States and Denmark.
However, MUUK was not an important part of the initial outbreak of scarlet fever in the UK in 2014. This was also not the strain involved in the outbreak of Strep A in the United Kingdom. Essex earlier this year.
More search
Professor Sriskandan said, "This type of strain has been around for a long time in developed countries 20 years ago, so the change we have seen in the UK may have more implications than normal." We do not know yet, it is too early to say if it will stay or if it will just die.
"Should we do more? I am a hospital doctor, so I see the much more serious side of the infectious disease caused by the bacteria … but it seems that the bacterial infections that cause sore throat seem to target the youngest children Children with scarlet fever are usually between 4 and 6 years old. We are also seeing an increase in strep throat around the same time of year. So I think we should look at whether a slightly more targeted diagnosis and the treatment of throat throat in this age group could reduce the reservoir of infection in the community.
"The much rarer infectious diseases, the invasive diseases that I see as a hospital doctor, are really very rare, but it is clear that if we could reduce the burden of strep throat and the Scarlet fever, it is very likely that we will reduce the burden of more invasive people. " serious infections too. "
She acknowledges that more research is needed: "We do not know if we conducted the study this year rather than 2016 if we would have found the same things, there is still work to be done."
"Plausible index"
In response to the findings of the Science Media Center, Professor Jimmy Whitworth, professor of international public health at the London School of Hygiene and Tropical Medicine, said: "This important study gives us a plausible indication of the recent worrying increase in cases of fever. Scarlet fever children in England. "
Researchers rightly ask for more surveillance to confirm these findings, as these streptococcal infections are highly susceptible to antibiotics, unlike most other types of sore throats in children. A vaccine, but perhaps more feasible in the short term, would be to re-evaluate and refine existing diagnostic tests that could greatly help general practitioners to accurately and quickly identify strep throat sore throat in the future . "
"The emergence of a dominant toxigenic clone M1T1 Streptococcus pyogenes during increased activity of scarlet fever in England: a population-based molecular epidemiological study" by Lynskey et al. The Lancet Infectious Diseases, Tuesday, September 10th. DOI: 10.1016 / S1473-3099 (19) 30446-3
[ad_2]
Source link