[ad_1]
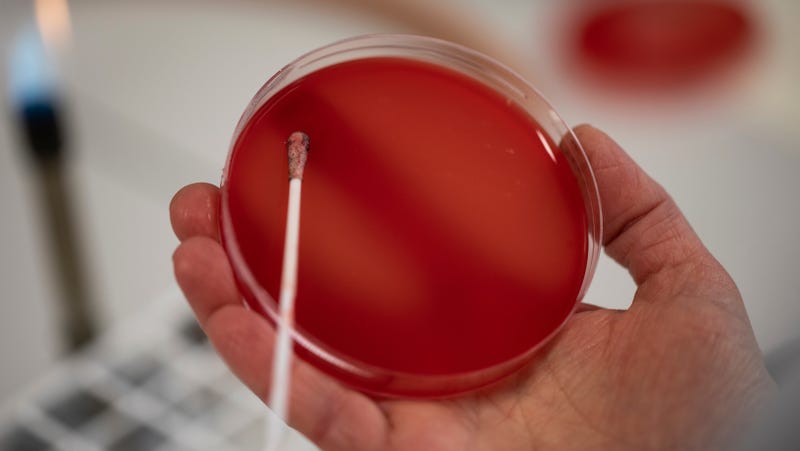
Here is a small piece of horror to transport you throughout the weekend. Scientists at North Carolina State University claim to have discovered causing the disease Salmonella bacteria in the United States able to resist one of the few antibiotics available for some infections. Worse still, the mutation responsible for this newfound resistance can easily be passed on to other bacteria, which increases the risk that some infections eventually become untreatable with antibiotics.
The last-resort antibiotic in question is a 50-year-old drug called colistin. At its peak, colistin was not the safest and most effective treatment. She ended up being badly received by the doctors, because it could cause serious lesions to the kidneys. But it has recently resurfaced as the last line of defense as it can still kill gram-negative bacteria resistant to multiple antibiotics. These bacteria include common insects such as Escherichia coli as well as hospital-acquired infections caused by Pseudomonas aeruginosa and Klebsiella pneumoniae.
Until now, there has been no evidence of widespread resistance to colistin among these specific superbugs. But scientists have begun to detect genetic mutations that allow bacteria to resist colistin from other bacteria – on free-motion DNA fragments called plasmids. After the initial release of the discovery of a mobilized colistin resistance gene (MCR) in 2015, at least two, or even at least eight other variants of the MCR have been found elsewhere.

A patient transplanted into feces was killed by a Superbug found in the stool of a donor
An aesthetically unpleasant but really promising medical treatment – a stool transplant – may come …
Read more
What makes this so dangerous is that plasmids can move between bacteria. It is therefore possible that these MCR genes could end up in a strain of E. coli resistant to all other drugs, for example. And if this strain infects and seriously ill a person, there may be no way imaginable for the doctors to treat it. If this event occurred again and again – and except for the discovery of a new or an old drug likely to occur in case of colistin failure – it would remain a reality where some infections, especially in hospitals or other areas where superbugs are prevalent, are simply impossible to treat.
It is this nightmare scenario that has forced physicians, including the authors of the latest study, to aggressively search for resistance genes, particularly RCM, in the wild.
According to this study, the discovery would have been discovered in an 18-year-old man suffering from food poisoning, including diarrhea, who had visited a doctor in 2014. After the tests, he was suspected of having fallen sick. MDR Salmonella. When the team tested her sample, she also found a variant of MCR, MCR3-1, on a plasmid.
Their work was published earlier in June in the Journal of Medical Microbiology.
Colistin is not usually used to treat Salmonellaalthough it is related to E. coli and other Gram-negative bacteria. And the man's infection could still be treated with other antibiotics. But the discovery of this particular combination of Salmonella bacteria and MCR 3.1 – the first known example, according to the researchers – is disturbing. Two weeks before his doctor's visit, he traveled to China. The widespread use of colistin in the country as an agricultural antibiotic, which only recently ended, would have accelerated the emergence of RCM. But as this case shows, MCR has long been internationalized, having been spotted in more than two dozen countries, both in humans and animals.
These cases of MCR are not impossible to treat (or even always to cause a disease) for the moment. But the more often they occur, the greater the chance that truly resistant and pan-resistant infections will become normal. Since this case really happened in 2014, it is also possible that the overall situation has only worsened since then.
All this to say: Have a good weekend!
[ad_2]
Source link