[ad_1]
DeepMind stunned the world of biology late last year when its AI model AlphaFold2 predicted the structure of proteins (a common and very difficult problem) so accurately that many claimed that the age-old problem of several decades was “solved”. Now researchers claim to have overtaken DeepMind in the same way that DeepMind has overtaken the rest of the world, with RoseTTAFold, a system that does almost the same thing at a fraction of the cost of computing. (Oh, and it’s free.)
AlphaFold2 has been talking about the industry since November, when it beat the competition at CASP14, a virtual competition between algorithms designed to predict the physical structure of a protein given the sequence of amino acids that compose it. DeepMind’s model was so far ahead of the rest, so precise and reliable, that many in the field were talking (half serious and in good spirits) of moving to a new area.
But one aspect that didn’t seem to satisfy anyone was DeepMind’s plans for the system. This has not been described in an exhaustive and open manner, and there were concerns that the company (which is owned by Alphabet / Google) was considering keeping the sauce more or less a secret to them – which would be their prerogative but also somewhat contrary to the ethics of mutual aid. in the scientific world.
Update: In a kind of surprise, DeepMind today published more detailed methods in the journal Nature. The code is available on GitHub. This greatly alleviates the aforementioned concern, but the advancement described below is still very relevant. I also added a comment from this team at the bottom of the post.
This concern seems to have been at least partly evoked by the work of researchers at the University of Washington led by David Baker and Minkyung Baek, published in the last issue of the journal Science. Baker, you may remember, recently won a Breakthrough Prize for his team’s work in combating COVID-19 with modified proteins.
The team’s new model, RoseTTAFold, makes predictions at similar levels of accuracy using methods that Baker, answering questions via email, frankly admitted, drawing inspiration from those used by AlphaFold2.
“The AlphaFold2 group presented several new high level concepts during the CASP14 meeting. From these ideas, and with a lot of collective brainstorming with colleagues in the group, Minkyung was able to make incredible progress in a very short period of time, ”he said. (“She’s amazing!” He added.)
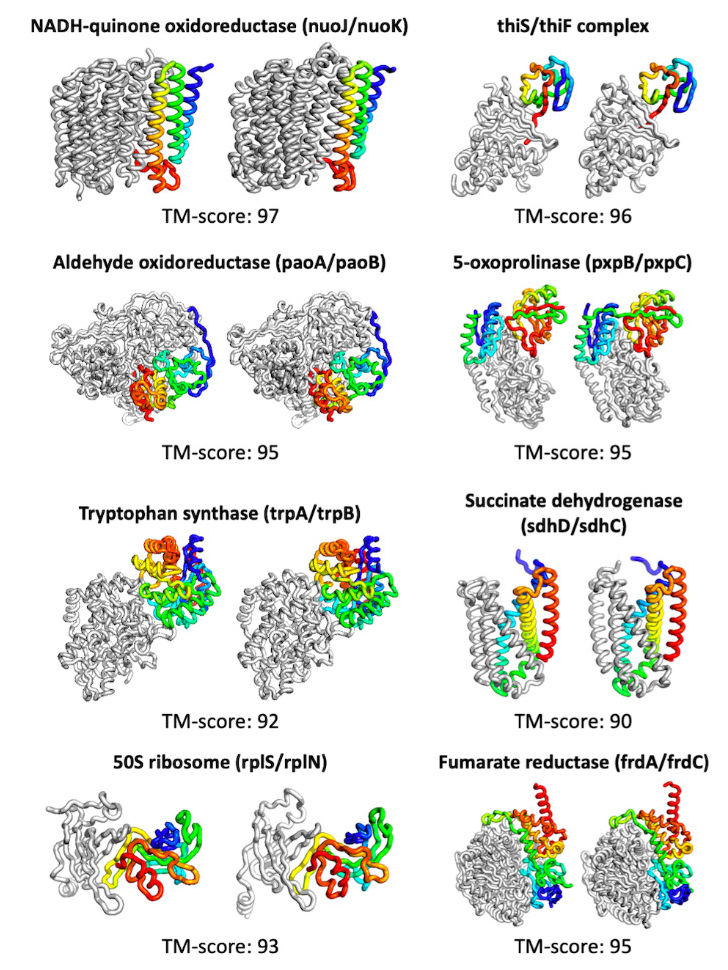
Examples of predicted protein structures and their basic truths. A score above 90 is considered extremely good. Image credits: UW / Baek et al
Baker’s group came in roughly second at CASP14, which is no small feat, but hearing DeepMind’s methods described has even usually put them on a collision course. They developed a “three-track” neural network that simultaneously considers the amino acid sequence (one dimension), the distances between residues (two dimensions) and the coordinates in space (three dimensions). The implementation is beyond complexity and well beyond the scope of this article, but the result is a model that achieves almost the same levels of precision – levels, it bears repeating, that were completely unprecedented less than 10 years ago. a year.
In addition, RoseTTAFold achieves this level of precision much faster, i.e. using less computing power. As the newspaper says:
DeepMind reported using multiple GPUs for days to make individual predictions, while our predictions are made in a single pass through the network in the same way that would be used for a server … the end-to-end version of RoseTTAFold requires about 10 minutes on an RTX2080 GPU to generate backbone coordinates for proteins with less than 400 residues.
Listen to this? It’s the sound of thousands of microbiologists sighing in relief and rejecting drafts of emails asking for the supercomputer’s time. It might not be easy to get your hands on a 2080 these days, but the point is that any high-end desktop GPU can do this task in minutes, instead of requiring a high cluster. range running for days.
The modest requirements make RoseTTAFold suitable for public hosting and distribution, something that might never have been in the cards for AlphaFold2.
“We have a public server to which anyone can submit protein sequences and have structures predicted,” Baker said. “There have been over 4,500 submissions since we installed the server a few weeks ago. We have also made the source code available for free.
It may sound very specialized, and it is, but protein folding has always been one of the most difficult problems in biology and a problem to which countless hours of high performance computing have been put into. You may remember Folding @ Home, the popular distributed computing application that allows people to donate their compute cycles in an attempt to predict protein structures. The kind of problem that could have taken thousands of days or weeks to resolve, essentially using brute-force solutions and checking adequacy, can now be fixed in minutes on a single desktop.
The physical structure of proteins is of the utmost importance in biology, as it is the proteins that perform the vast majority of the tasks in our body and the proteins that need to be changed, removed, improved, etc. for therapeutic reasons; first, however, they must be understood, and until November this understanding could not be reliably achieved by calculation. At CASP14 it was proven to be possible, and now it is widely available.
This is not, by far, a “solution” to the protein folding problem, although the sentiment has been voiced. The structure of most proteins at rest under neutral conditions can now be predicted, which has huge repercussions in multiple domains, but proteins are rarely found “at rest under neutral conditions”. They twist and contort to grab or release other molecules, to block or slip through doors and other proteins, and generally to do whatever they do. These interactions are far more numerous, complex, and difficult to predict, and neither AlphaFold2 nor RoseTTAFold can.
“There are many exciting chapters to come… the story is just beginning,” Baker said.
Regarding the DeepMind article, Baker made the following comment in a spirit of collegial camaraderie:
I have read and think this is a nice article describing a fantastic job.
The DeepMind paper is actually very complementary to our paper, and I think it’s appropriate that it doesn’t come out after ours, because our work is really based on their advancements.
I think readers will enjoy reading both articles – they are far from redundant. As we point out in our article, their method is more accurate than ours, and now it will be very interesting to see which features of their approach are responsible for the remaining differences. We are already using RoseTTAFold for protein design and more systematic prediction of protein-protein complex structure, and we are excited to improve them quickly, as well as traditional single-chain modeling, incorporating ideas from the article by DeepMind.
If you are curious about the science and the potential repercussions, consider reading this much more detailed and technical account of the methods and possible next steps written as a result of the CASP14 performance of AlphaFold2.
[ad_2]
Source link