[ad_1]
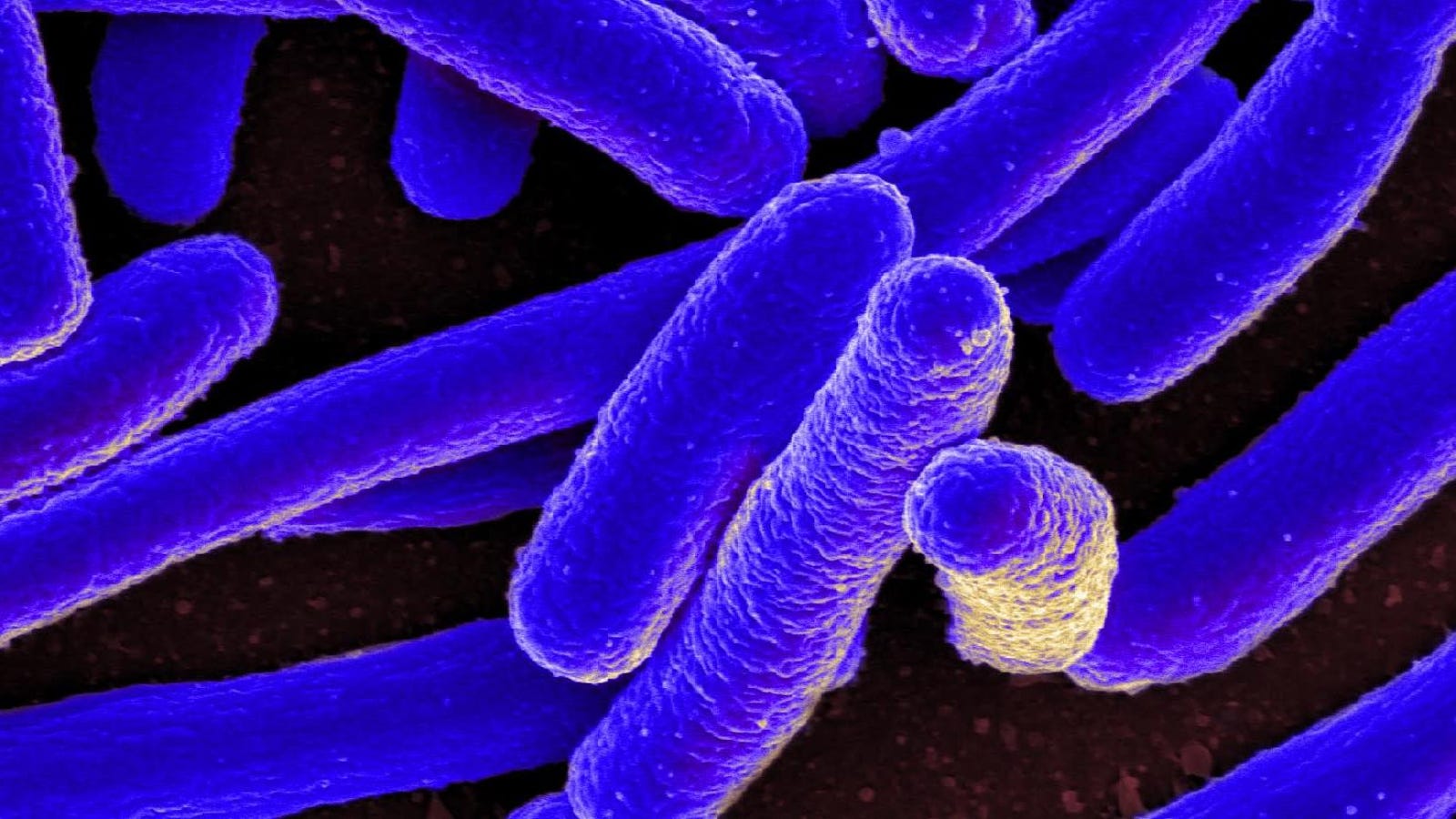
Scientists have created a variant of the E. coli bacteria with a fully synthetic genome, according to a new paper.
However, building and replacing the entire large genome was only one goal of the team at the Molecular Biology Laboratory of the Medical Research Council in Cambridge, UK. They hoped that the resulting bacteria would use a reduced number of possible combinations of base pairs of DNA to produce the 20 amino acids. In the future, the now obsolete sequences could be used to produce proteins and amino acids never seen before.
The purpose of the document was not simply to reconstruct the genetic code of a bacterium, but to simplify redundancies in order to have more genetic code on which to work to create custom genomes. The genetic code is written in four letters: A, T, C and G, which represent the molecules adenine, thymine, cytosine and guanine. These nucleotides can form 64 three-letter codons, most of which correspond to an amino acid, the building blocks of the proteins that make life work. All but two amino acids are encoded by several synonymous codons. The researchers wanted to see if they could rewrite the E. coli The genome of the bacterium with fewer codons, like rewrite the dictionary but representing all the sounds "k" hard with only the letter k, instead of using sometimes c or q like English.
The production of the genome required a first drafting. The researchers devised a genome in which they replaced two codons that encode the amino acid serine by synonyms, and did the same with the stop codon, which tells cell functions to stop reading a strand of DNA during the construction of a protein. Next, the researchers built their DNA using the different laboratory techniques already used in synthetic biology. Finally, they had to replace the genetic material of the bacterium with synthetic DNA. They could not just transfer everything: they had to break their genome into pieces, transplanting them into living bacteria little by little until they replaced the whole body. E. coli genome, according to the article published in Nature.
the E. coli according to the New York Times, they survived, but they slowed down and lasted longer. But the new E. coli cells rely on only 61, rather than 64 codons in total.
It was a tour de force, said a scientist in Gizmodo. "For those of us working in synthetic genomics, this is the most exciting title; they synthesized, built and showed that a synthetic genome of 4 million base pairs could work, "Gizmodo told Tom Ellis, director of the Synthetic Biology Center at Imperial College London, who had examined the document. "It's more than anyone before."
Sci-fi speculation aside, there are several potential uses for a synthetic genome like this one. There are obvious biotechnological applications: the elimination of redundancies allows researchers to have additional codons, possibly to develop new amino acids, proteins and bacteria capable of doing new things. The codon swap also serves as a firewall against viruses that can turn the cell around, Ellis said. It's a rapidly evolving field, and it's one of many groups doing this kind of genome permutation.
The methods that researchers have used to swap the genome provide "a blueprint for future genome synthesis," according to the article. And then, the researchers hope to understand how much they could further rationalize. E. coli genome, as well as what they can do with the codons they released.
[ad_2]
Source link