[ad_1]
Over the past decade, decreasing costs of chemical synthesis of DNA and improved methods of assembling DNA fragments have allowed researchers to extend the synthetic biology in the generation of chromosomes and whole genomes. Until now, synthetic DNA has been constructed with one million base pairs, including a set of yeast chromosomes. Saccharomyces cerevisiae and several versions of the genome of the bacteria Mycoplasma Mycoides1,2. Now, by writing NatureFredens et al.3 signal the completion of a synthetic version of 4 million base pairs of the Escherichia coli genome. It is a landmark in the emerging field of synthetic genomics and is finally applying this technology to the laboratory's most successful bacterium.
Synthetic genomics offers a new way of understanding the rules of life, while evolving synthetic biology into a future in which genomes can be written for design. The pioneers of the field – researchers at the J. Craig Venter Institute in Rockville, Maryland – have used this method to better define the minimal set of genes required for a free cell. By adopting an approach that consists of redefining segments of the genome by computer, chemically synthesizing fragments and then assembling them, these pioneers have succeeded2 by reducing the size of the M. mycoides genome of about 50%. Doing the same thing with only genome editing tools would be much more laborious, like working with E. coli demonstrates: here, gene deletion methods have eliminated, at best, only 15% of the genome4.
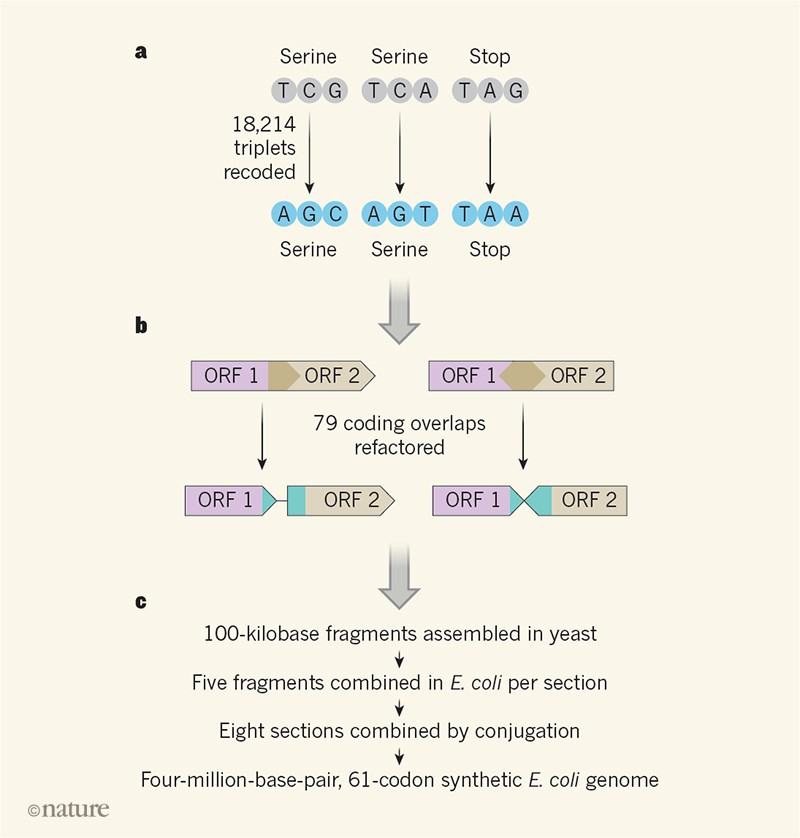
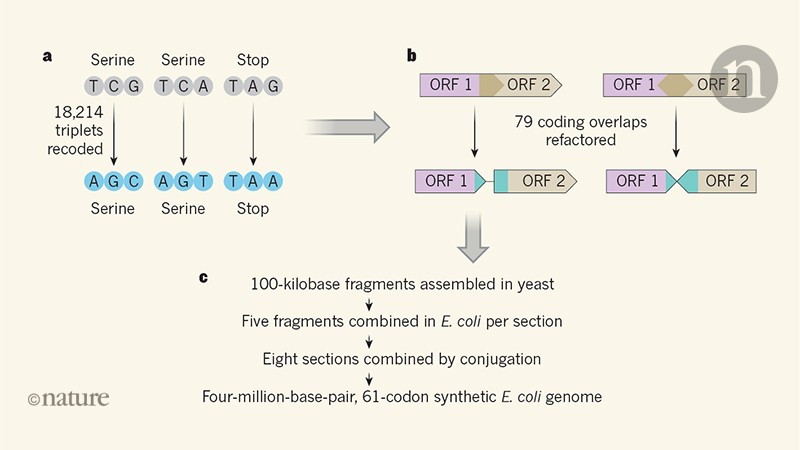
Fredens and his colleagues used this reduced genome of E. coli as a model for a synthetic genome with another type of minimization in mind – codon reduction. The genetic code has an inherent redundancy: there are 64 codons (triplets of "letters" or bases) to encode only 20 amino acids plus the "start" and "stop" points that mark the beginning and end of a stretch protein coding. sequence. This redundancy means, for example, that there are six codons coding for serine amino acids and three possible stop codons. Through design, synthesis and assembly, Fredens et al.3 were able to build a E. coli genome that uses only 61 of the 64 available codons in its protein coding sequences, replacing two serine codons and a stop codon with synonyms (the codons are "spelled" differently but give the same instruction). Previous works using genome editing tools have already produced a synthesis E. coli which uses only 63 of the 64 codons, but this required that the stop codons with the TAG sequence (there were only 321 around the genome) be replaced by another stop codon5. The 61 codon reduction required modification of 18,214 codons, requiring a genome synthesis approach.
Fredens and his colleagues built their synthetic E. coli genome using large-scale DNA assembly methods and genome integration that they had previously developed6 to probe the limits of codon changes in E. coli. In their approach (Fig. 1), DNA is computer-designed, synthesized chemically and assembled into 100 kilobase fragments in vectors. S. cerevisiae; these vectors are then taken over by E. coli and integrated into the genome in the direct place of the equivalent natural region. By iterating this process five times, DNA sections of 500 kilobases have been replaced by synthetic versions. Eight strains of E. coli were produced in this manner, each harboring sections of synthetic DNA that covered a different region of the genome. These sections were then combined using conjugation methods to form the complete synthetic genome.
Large-scale construction has been an impressive success, with very low transfer rates off target, but has not proceeded smoothly. Many genes in the E. coli the genome partially overlaps with others, and in 91 cases the overlapping regions contained codons that needed to be changed. This is complex because synonymous modifications in a protein coding sequence can alter the amino acids encoded by the overlapping one. To tackle this, the team "refactored" 79 sites in the genome, duplicating the sequence to separate the superimposed coding sequences into individual coded sequences (Fig. 1). While this approach has generally been successful, it has required careful debugging in a few cases where refactoring has also altered gene regulation.
The final strain proved viable and was able to grow under a variety of laboratory conditions, although it is a little less vigorous than its natural counterpart. It no longer uses the TAG stop codon or the two TCG and TCA serine codons, so that the cellular machinery that recognizes them can now be deleted or reassigned to recruit "non-canonical" amino acids beyond 20 usually used by most living cells. This recruitment has already proved useful in codon codon 63. E. colifor both biotechnology projects in which non-canonical amino acids are encoded in the desired sequence positions to provide residues that can participate in chemical reactions that natural proteins can not; and for biosecurity reasons, in that the natural transfer of legible information encoded by DNA inside and outside the E. coli is limited because the cell works with a genetic code slightly different from the rest of the natural world5. Expect all these applications to be extended in the new codon 61 E. coli, which could code the use of more than one non-canonical amino acid and generate a stricter genetic firewall (because 3 of the 64 codons are no longer recognized).
The synthesis of a 4 million base pair genome and the reduction of the 61 codon genetic code are new registrations for synthetic genomics, but may not last much longer. The international Sc2.0 consortium is approaching the synthesis of the 16 chromosomes of the 12 million base pair S. cerevisiae genome – the first synthetic genome of a eukaryotic organism, the group that includes plants, animals and fungi – and the synthesis of a codon 57 E. coli the genome is also underway1,7. A genome of the bacteria Salmonella Typhimurium that has two fewer codons than the natural body is also under construction8. This could one day allow bacteria with a synthetic genome to be used as cell-based technologies in the human gut.
From a technological point of view, the most interesting aspect of all these projects is that the synthetic genome construction processes are remarkably similar, with sections in kilobases of synthesized DNA being assembled (by the way). homologous recombination process) in 50 to 100 kilobases. pieces in yeast cells, these pieces then being used to replace natural sequences within the target organism (by selectable recombination methods). The standardization of methods will automate the steps and allow more research groups to enter the field. Genome minimization and codon reduction are only the first uses of this new technology, which could someday give us functionally reorganized genomes and tailored genomes to direct cells to specialized tasks.
Sign up for the everyday Nature Briefing email
Stay abreast of what matters in science and why, chosen by hand Nature and other publications around the world.
S & # 39; register
[ad_2]
Source link