[ad_1]
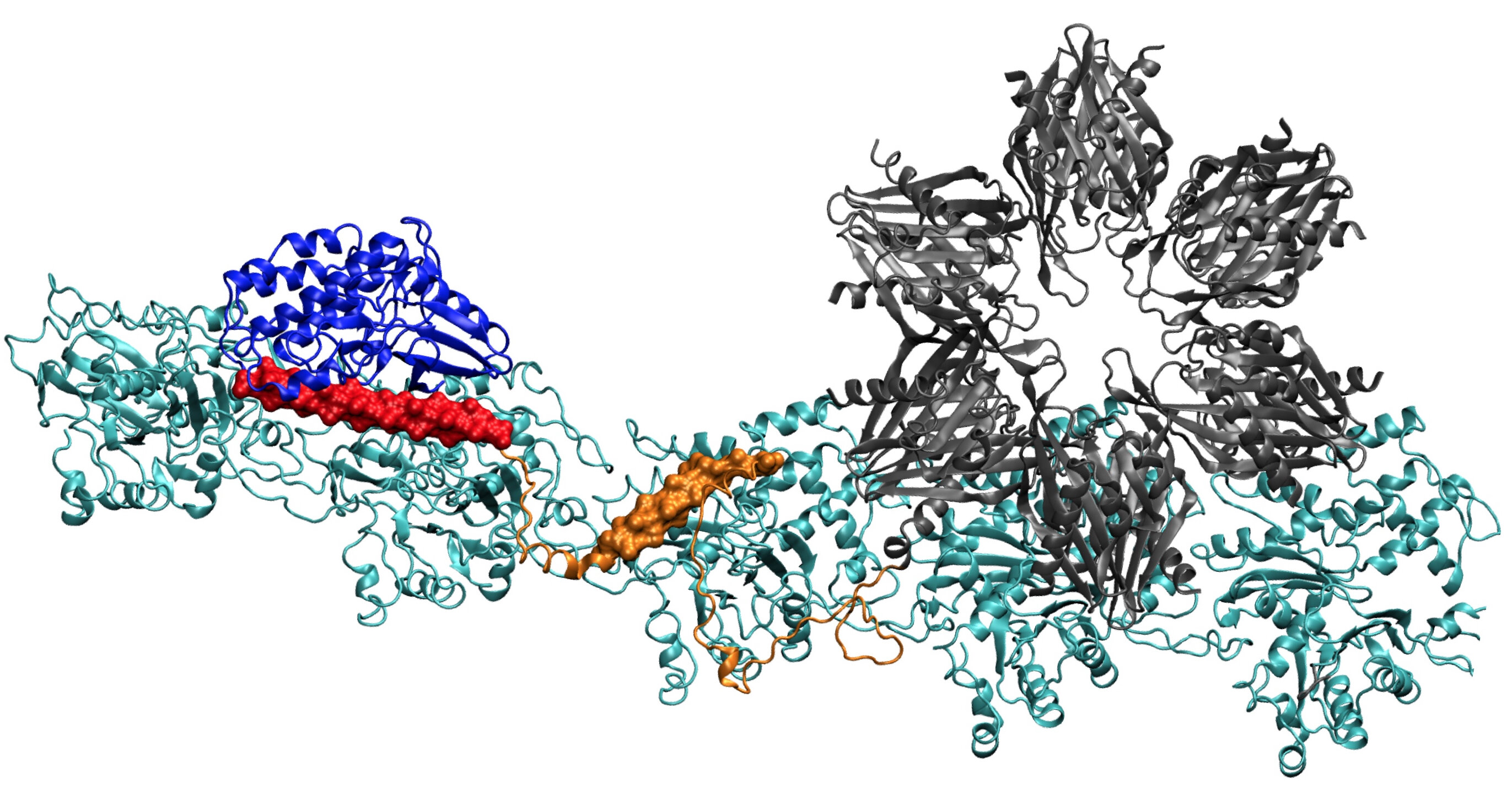
A model of the CaMKII protein shows several domains that allow it to bind actin filaments in dendrites of neurons into bundles, thus giving shape to dendrites. Researchers from Rice University, the University of Houston and the Health Sciences Center of the University of Texas at Houston believe that this complex is essential for long-term memory formation. term. Credit: Wolynes Research Laboratory / Rice University
A new piece of a difficult puzzle – the nature of memory – is set up this week with a hint on how brain cells change their structure when they learn something.
Interactions between three moving parts – a binding protein, a structural protein, and calcium – are part of the process by which electrical signals enter neural cells and alter the molecular structures that are supposed to enable memory cognition and storage.
Colleagues from Rice University, the University of Houston (UH) and the University of Texas Health Sciences Center in Houston (UTHealth) have combined theories, simulations and experiments to determine how a central binding protein, calcium-calmodulin-dependent kinase II (CaMKII) – binds and detaches from the cytoskeleton of a neuron.
The report of the team in the Proceedings of the National Academy of Sciences gives the first clear details of how CaMKII binding sites act to align actin filaments – the structural protein – into long, rigid bundles. The bundles serve as skeletons to dendritic spines, spiky protuberances that receive chemical messages through synapses of other neurons.
Peter Wolynes, a theoretical physicist at Rice, joins the ongoing collaboration of physicist Margaret Cheung and neurobiologist UTHealth Neal Waxham, who aimed to understand how signals cross dendrites, the branches of nerve cells that transmit information between cells.
Finding the complete structure of CaMKII has proven to be too complex for X-ray crystallography, although some parts of its structure are known. When combined with the actin that makes up the cytoskeleton, the system also becomes the largest protein that Wolynes and his team have analyzed through its protein structure prediction program, AWSEM.
Once done, the computer-predicted structure matched remarkably well with Waxham's and his group's two-dimensional electron microscope images, which clearly show that the parallel actin filaments are held together, like a scale, by bars of CaMKII.
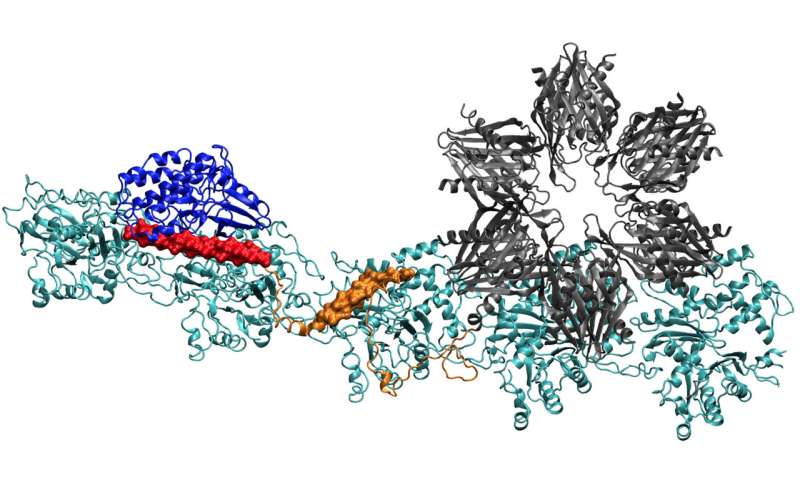
The CaMKII protein at the top of the docks in three pockets on actin filaments twisted into neurons, a process that researchers at Rice University, the University of Houston, and the Science Center of the United States. the health of the University of Texas at Houston (UTHealth) consider important for the formation of long-term memory. The regulation domain (in red) of CaMKII also binds to incoming calmodulin proteins, which decompress the entire actin structure and allow the clustered filaments to rearrange. Credit: Wolynes Research Laboratory / Rice University
"There are certainly preliminary chemical steps involving the enzyme activity of CaMKII before reaching this stage; therefore, we do not have a very clear idea of how to put everything in place. "said Wolynes. "But it's clear that the assembly of the complex is the key step where chemistry turns into a larger-scale structure that can hold a memory."
CaMKII is particularly well suited to interact with actin, the most abundant protein in eukaryotic cells and which has special abilities in neurons, where it must not only give rise to thousands of dendrites (in each of billions of neurons) their form of rest, but also give them a level of plasticity to adapt to a constant barrage of signals.
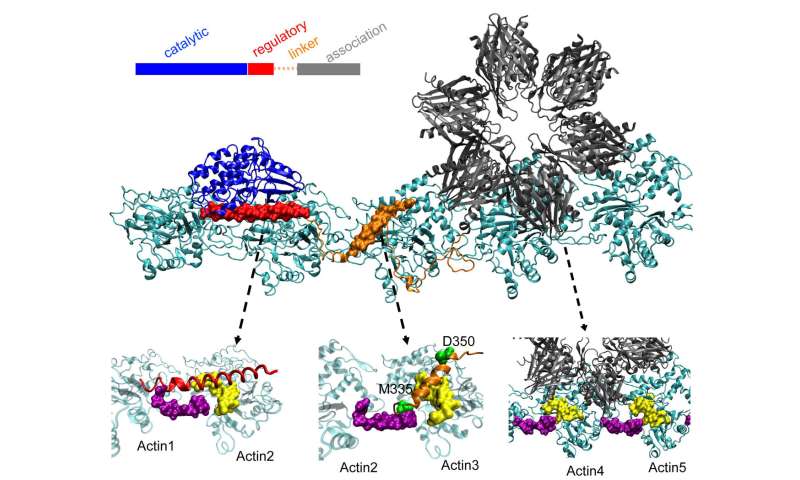
Actin molecules self-assemble into long, twisted filaments. The hydrophobic pockets between these molecules are perfectly configured to bind to CaMKII, a large protein with multiple parts or domains. These domains latch on three consecutive binding sites on the filament, and twists place binding sites at regular intervals to prevent proteins from piling up.
The "association" domain of CaMKII is a six-fold subunit that also binds to adjacent filaments to form actin bundles, the backbone of the dendritic spines that give shape to these protuberances.
These bundles remain rigid if the dendrite contains little calcium. But when the calcium ions enter the synapse, they combine with the calmodulin proteins, allowing them to bind to another part of CaMKII, the regulatory domain of the floppy disk. This triggers the dissociation of a CaMKII domain from the filament, followed by the rest of the protein, opening a short window of time during which the beams can reconfigure themselves.
"When the amount of calcium provided is sufficient, activated calmodulin destroys these structures, but only for a while," said Wolynes. "Then, the cytoskeleton is reforming, while the dendritic spine can take on a different shape that may be larger."
"We know that calcium brings information to the cell," added Cheung. "But how nerve cells know what to do with it really depends on how this protein codes information." Part of our job is to link that to the molecular level and then project how these simple geometric rules develop microscopic structures bigger."
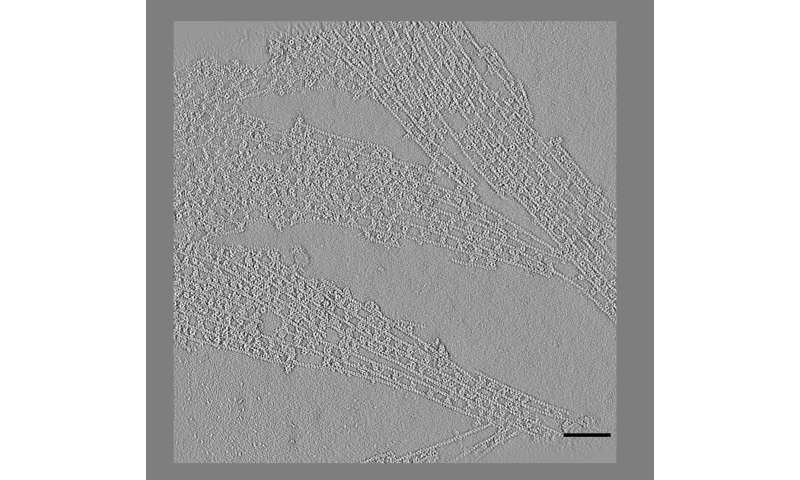
An electron microscope image shows actin filaments held together in parallel or branched arrangements by CaMKII proteins in a neuron. Simulations and experiments conducted at Rice University, the University of Houston and the University of Texas Health Sciences Center in Houston have shown that the spacing between Adjacent particles of CaMKII in the complexes is almost fixed at 36 nanometers. The scale bar is 100 nanometers. The image was taken at the Structural Biology Imaging Center at McGovern School of Medicine at UTHealth. Credit: Waxham Lab / UTHealth
The team's calculations showed that the field of association is responsible for about 40% of the binding force of the protein to actin. A linking domain adds another 40% and the crucial regulatory area provides the final 20%, a sensible strategy, as the regulatory field is on the lookout for incoming calcium calmos which can extract all of the protein from the filament.
The project originated from Rice's Center for Theoretical Biological Physics (CTBP), of which Wolynes is the co-director and Cheung, a senior scientist. Their association dates back to the time when they were both at the University of California at San Diego. He was a professor and studied at the graduate level of physicist Rice, José Onuchic, also co-director of CTBP. Wolynes has also served on her thesis review committee, she said.
Cheung was aware of earlier work by Wolynes and his group Rice, who suggested that actin stabilizes the prionic fibers that are supposed to encode memories in neurons, and that his research with Waxham went hand in hand with the role of calcium. in the activation of CaMKII.
"This is one of the most interesting problems in neuroscience: how do short-term chemical changes lead to something long, like memory?" Said Waxham. "I think one of the most interesting contributions we bring is to explain how the system takes into account the changes that occur in milliseconds and creates something that can survive the initial signal."
The puzzle is far from over, said Wolynes. "Margaret and Neal's earlier work involved the initiation of memory events," he said of his colleagues' study of calmodulin. "Our article on prions focused on preserving memory, at the end of the learning process, and actin is at the center, there can be many other things in the middle."
"These big issues are of interest to many people," he said. "It's a key part of the problem, but it's clearly not the end of the story."
Unexpected mechanism allows protein kinase to decode calcium signaling in the brain
Qian Wang et al., "Calcium-calmodulin-dependent kinase II – Actin assemblages and their dynamic regulation by calmodulin in dendritic spines," PNAS (2019). www.pnas.org/cgi/doi/10.1073/pnas.1911452116
Quote:
Scientists advance in the search for the molecular roots of memory (August 26, 2019)
recovered on August 26, 2019
at https://phys.org/news/2019-08-scientists-advance-memory-molecular-roots.html
This document is subject to copyright. Apart from any fair use for study or private research purposes, no
part may be reproduced without written permission. Content is provided for information only.
[ad_2]
Source link