[ad_1]
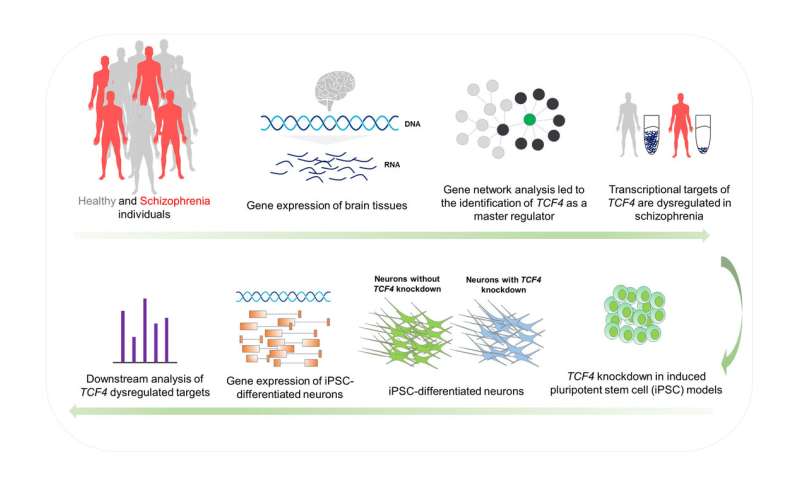
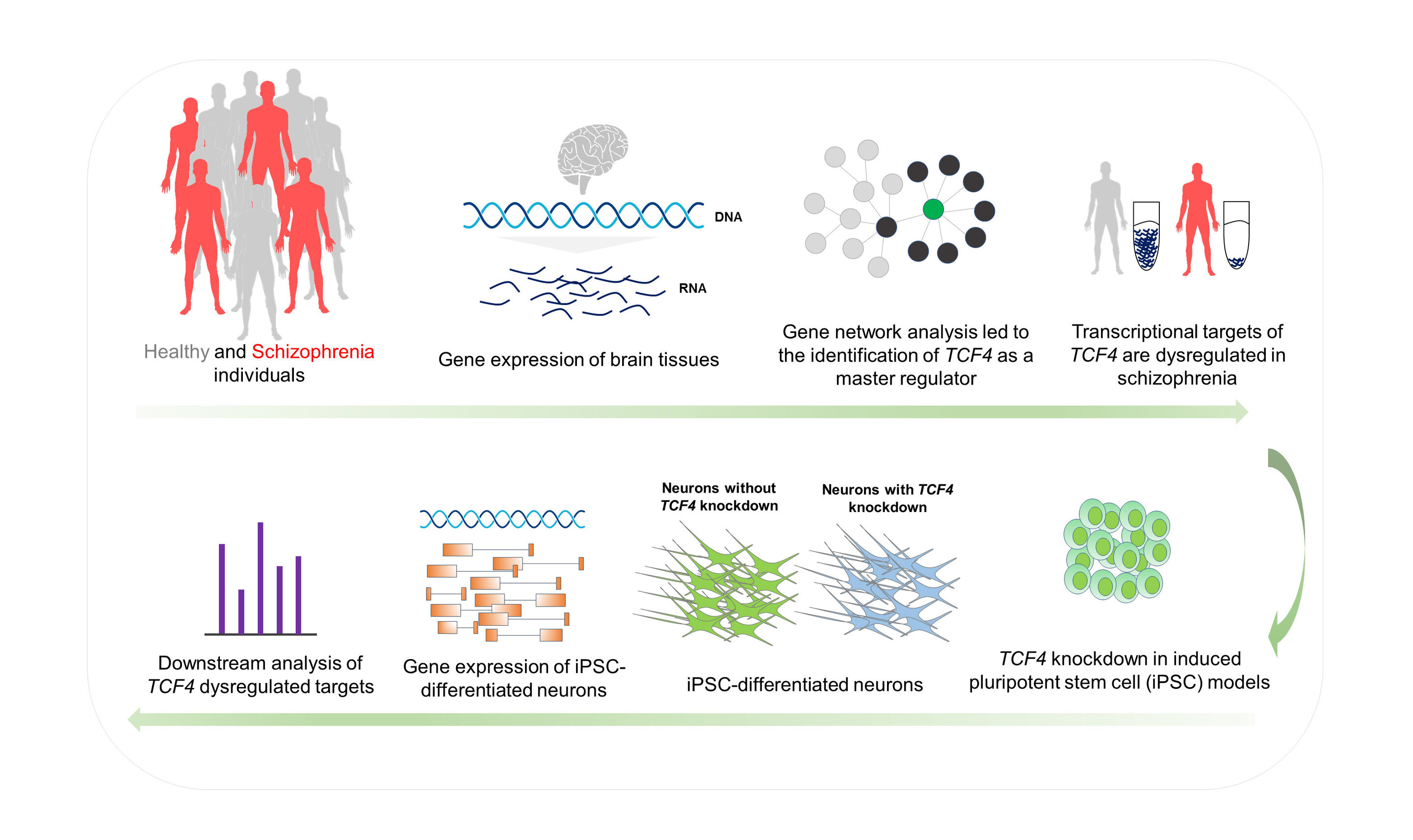
Illustration of the study plan to identify and validate TCF4 as a key gene for schizophrenia. Credit: Abolfazl Doostparast Torshizi, Jubao Duan and Kai Wang
Using computer tools to study gene transcription networks in large collections of brain tissue, a scientific team has identified a gene that plays the role of a major regulator of schizophrenia during the early development of the human brain. The results could lay the foundation for future treatments of the highly complex neuropsychiatric disorder.
"Because hundreds or even thousands of genes can contribute to the risk of schizophrenia, it is essential to understand which are the most important, orchestrating the central networks of the disease," said study director, Kai Wang, Ph.D. the Department of Pathology and Laboratory Medicine and the Raymond G. Perelman Center for Cellular and Molecular Therapy (CCMT) of the Children's Hospital of Philadelphia (CHOP). "Identifying key regulators can help us focus on priority targets for new treatments in the future."
The study was put online today in Progress of science. Wang co-directed the research with Jubao Duan, Ph.D., Charles R. Walgreen Research Chair and Associate Professor at the North Shore University's HealthSystem Psychiatric Genetics Center, and University of Chicago, Evanston, Ill. The first author was Abolfazi Doostparast Torshizi, Ph.D., from CCMT at CHOP.
Although schizophrenia affects about 1 in 100 adults and is highly hereditary, the genetic architecture of the neuropsychiatric disorder is notoriously complex and has many unresolved issues. Wang and his colleagues compare the current state of knowledge to recent advances in cancer research. Oncology researchers have identified many gene pathways and networks that, when disturbed, result in specific types of cancer. This knowledge has made fine distinctions in the diagnosis of subsets of cancer types and led to individualized treatments based on the patient's genetic profile.
The underlying genomic biology in neuropsychiatric disorders is probably more difficult. In the most extreme case, researchers have recently proposed an "omnigenic" model, in which almost all the genes of a cell type involved in the disorder contribute to a specific neuropsychiatric disorder, but Wang notes that "all genes do not have the same weight – the problem is to determine which ones are more important than others. "
Thus, the research team used computer systems biology approaches to discern a central pathway of schizophrenia relevant to the disease and to discover a major regulator in this pathway that affects hundreds of downstream genes.
Wang and colleagues analyzed two sets of independent data from biological samples of patients with schizophrenia and control subjects. A dataset, the CommonMind Consortium (CMC), is a public-private partnership with well-organized brain collections. The other was a collection of primary cultured neuronal cells derived from the olfactory epithelium (CNON), generated by co-authors of the study at the University of Southern California and at SUNY Downstate. The CMC dataset contained postmortem brain tissue in the adult, while the CNON dataset, used to validate the results of the CMC study, represented cultures of cells containing neuronal cells derived from nasal biopsies. Applying an algorithm developed at Columbia University to reconstruct gene transcription networks, the research team identified the gene TCF4 as the main regulator for schizophrenia.
Previous genome-wide association studies (GWAS) had shown that TCF4 was a place for the risk of schizophrenia, said Wang, but the functional effects of the gene were poorly understood. The research team investigated these effects by inhibiting or decreasing gene expression in neural progenitor cells and glutamatergic neurons derived from pluripotent stem cells induced in the Duan laboratory at NorthShore.
Observations on three different cell lines showed that, once reversed, the predicted value TCF4 Regulatory networks have been enriched for genes with transcriptomic modifications, as well as for genes involved in neuronal activity, genes at risk for schizophrenia with genomic significance, and de novo mutations associated with schizophrenia. Although some of the cellular effects of TCF4 dysregulation have already been shown in mice, Duan noted that the results of the disturbance TCF4 Gene networks in human stem cell models may be more relevant to the neurodevelopmental aspects of neuropsychiatric disorders.
The present study, said the researchers, paves the way for new investigations. One of the directions, said Wang, is to use extended data sets to determine if other key regulators, in addition to TCF4 can act in schizophrenia. If this is the case, it may be possible to classify patients with schizophrenia into subgroups that are more sensitive to specific treatments, as is the case in many cancers, in order to facilitate the implementation of schizophrenia. a precision medicine in psychiatric diseases.
Doostparast added that other approaches could involve the pursuit of functional genomics in individual cells to evaluate which cell types are most influenced by dysregulatory gene expression.
The study represents one of the first successful examples of combining computer approaches and experimental models based on stem cells to unravel complex gene networks in psychiatric diseases.
Wang noted the limitations of the study. Empirical validation has focused on neural progenitor cells and glutamatergic neurons, but other types of cells could be explored in the future, such as interneurons and microglia, which have also been involved in the development of schizophrenia.
The neurobiological mechanisms underlying schizophrenia may depend on sex
A. Doostparast Torshizi et al., "Deconvolution of transcription networks identifies TCF4 as the main regulator in schizophrenia," Progress of science (2019). DOI: 10.1126 / sciadv.aau4139, https://advances.sciencemag.org/content/5/9/eaau4139
Quote:
Scientists identify gene as major regulator in schizophrenia (September 11, 2019)
recovered on September 11, 2019
on https://medicalxpress.com/news/2019-09-scientists-gene-master-schizophrenia.html
This document is subject to copyright. Apart from any fair use for study or private research purposes, no
part may be reproduced without written permission. Content is provided for information only.
[ad_2]
Source link